Abstract
The gene DTNBP1 encodes the protein dysbindin and is among the most promising and highly investigated schizophrenia-risk genes. Accumulating evidence suggests that dysbindin plays an important role in the regulation of neuroplasticity. Dysbindin was reported to be a stable component of BLOC-1 complex in the cytosol. However, little is known about the endogenous dysbindin-containing complex in the brain synaptosome. In this study, we investigated the associated proteome of dysbindin in the P2 synaptosome fraction of mouse brain. Our data suggest that dysbindin has three isoforms associating with different complexes in the P2 fraction of mouse brain. To facilitate immunopurification, BAC transgenic mice expressing a tagged dysbindin were generated, and 47 putative dysbindin-associated proteins, including all components of BLOC-1, were identified by mass spectrometry in the dysbindin-containing complex purified from P2. The interactions of several selected candidates, including WDR11, FAM91A1, snapin, muted, pallidin, and two proteasome subunits, PSMD9 and PSMA4, were verified by coimmunoprecipitation. The specific proteasomal activity is significantly reduced in the P2 fraction of the brains of the dysbindin-null mutant (sandy) mice. Our data suggest that dysbindin is functionally interrelated to the ubiquitin-proteasome system and offer a molecular repertoire for future study of dysbindin functional networks in brain.
Keywords: Dysbindin, schizophrenia, synaptosome, protein complex, protein−protein interaction, BAC transgenesis, proteasome, mass spectrometry
Introduction
Schizophrenia (SCZ) is a chronic and severe mental disorder affecting ∼1% of people in their lifetime.1 Epidemiological studies indicate that genetic risk factors are important to its causation because heritability was estimated up to ∼80% based on family and twin studies.1−4 Many putative SCZ susceptibility genes have been identified by studies such as genomic sequence analyses of single nucleotide polymorphisms (SNPs), haplotypes, and copy number variants in individuals with and without SCZ.5−9 The accumulating evidence suggests that the genetic factors of SCZ are very complex and polygenic. Multiple variants with moderate effects in different genes or loci (interacting with each other and environmental factors) are likely to contribute to the etiology.8−10 The combinational variants may affect multiple biological pathways, disturbing signal transduction, molecular function, and neuronal connectivity in the brain. In agreement with this hypothesis, post-mortem and brain image studies on SCZ patients have revealed that abnormalities of synaptic connectivity are a hallmark feature of SCZ.11
The dystrobrevin-binding protein 1 (DTNBP1) gene is among the most promising and highly investigated SCZ-risk genes.12−14 Despite some inconsistent findings,15−17 numerous association studies have reported linkage between the risk variants (SNPs or haplotypes) in the DTNBP1 gene and the occurrence and symptom severity of SCZ.7,18−20 The involvement of DTNBP1 in SCZ is also supported by the observations in which both the transcript and protein levels of DTNBP1 were reduced in the hippocampus and the dorsolateral prefrontal cortex of SCZ patients.21−24 While none of the risk variants were found to change protein amino acid sequence, it was proposed that these risk variants could act as cis-elements to decrease DTNBP1 expression.22,25 Moreover, the studies on Dtnbp1-null mutant mice (also called sandy or sdy mice), which exhibit SCZ-related behaviors such as social withdrawal and cognitive deficits,26−28 also support the role of DTNBP1 as a SCZ risk gene.
DTNBP1 encodes a coiled-coil-containing protein family commonly called dysbindin or dysbindin-1, which was identified initially as an associated protein of α- and β-dystrobrevin in the muscle and brain of mice.29 Dysbindin is ubiquitously expressed in neurons of brain areas such as hippocampus, striatum, and cortex and is enriched in synapses.3,11,30 DTNBP1 expresses a group of alternative spliced transcripts. There are 16 alternative spliced transcripts of DTNBP1 identified in humans, but only three isoform proteins, dysbindin-1A, -1B, and -1C, could be detected.24,31 Interestingly, these isoforms appear to have different functions, as they have distinct subsynaptic localization in pre- and/or postsynaptic regions.23 The expression of dysbindin in mouse brain was found to be developmentally regulated. The level of dysbindin is higher in embryonic stages and gradually declines until adulthood, suggesting its potential function in early development.32 Studies using model animals show increasing evidence that dysbindin plays a role in the regulation of neuroplasticity. In particular, abnormalities of glutamatergic, dopaminergic, and GABAergic transmission were observed in sandy mice.11 As the reduction of dysbindin is also found in the synaptic region of SCZ patients, these data suggest that dysbindin contributes to impaired synaptic transmission in SCZ.21,23,33 In addition to SCZ, the mutation of DTNBP1 is also associated with Hermansky–Pudlak syndrome and bipolar disorder.34,35
As the genetics of SCZ are polygenic and appear to involve multiple genes and signaling pathways,9 understanding the biology of putative risk genes and their associated functional networks is critical to shed the light on the obscure pathogenesis and the development of new therapeutics. The study of protein–protein interactions (PPIs) has become one of the essential tools to decipher biological processes and functional networks.36 The interactome study using yeast-two hybrid system on dysbindin and disrupted in schizophrenia 1 (DISC1), another promising and extensively studied SCZ risk gene, revealed their involvement in common biological processes, as common PPIs were identified from their interactomes.37 To date, more than 140 dysbindin-associated proteins have been reported, but only few of the PPIs have been confirmed in the brain.12 Many of the PPI candidates were acquired from non-neuronal tissues (i.e., liver) or cell lines (i.e., HEK293 and SH-SY5Y), and the proteomes associated with dysbindin in non-neuronal tissues and cell lines could be different from that in brain.12,34,38,39 In addition, although dysbindin is a soluble protein and is present largely in the soluble cytosol of the brain, it is also localized to the synaptosome, the insoluble membrane structure containing the molecular machinery essential for synaptic transmission.30,32 A gel filtration study based on complex size and cofractionation of the BLOC-1 subunit pallidin suggested that dysbindin is present in the BLOC-1 complex in the cytosol of brain,32 but little is known about the dysbindin-associated complexes in the synaptosome, where dysbindin may exert its functional role in the regulation of neuroplasticity. Moreover, affinity purification of target proteins under the intrinsic conditions of organisms using specific antibodies or tags (i.e., tandem affinity purification) followed by mass spectrometric analysis is particularly useful to identify protein complexes that exist in vivo.40 However, this type of systematic study has not yet been reported for the dysbindin complexes in the brain.
In this study, we investigated the associated proteome of dysbindin in the P2 synaptosome fraction from mouse brain.41 We first profiled dysbindin-containing complex in the P2 fraction using chemical cross-linking and sucrose density gradient ultracentrifugation. Our data suggest that dysbindin has at least three isoforms associating with different complexes in the P2 fraction of mouse brain. The largest dysbindin (dysbindin-1A) is present in complex(es) larger than that of BLOC-1. We next generated BAC transgenic mice expressing a tagged dysbindin and purified dysbindin-containing complexes from the P2 fraction using immunopurification. Forty seven putative dysbindin-associated proteins were identified by mass spectrometry, including all known components of BLOC-1. We used pathway analysis to guide recognition of noncanonical functional networks of the identified proteins. A selected set of candidates were verified by coimmunoprecipitation (coIP), including WDR11, FAM91A1, snapin, muted, pallidin, and two of the three identified proteasome subunits, PSMD9 and PSMA4. On the basis of these observations, we performed proteasome activity assays and found that specific proteasomal activity is significantly reduced in the P2 extract of sandy mice.
Materials and Methods
Antibodies
The following antibodies were obtained commercially: anti-V5 (Invitrogen, R960), anti-β-tubulin (Developmental Studies Hybridoma Bank, E7), anti-FLAG M2 (Sigma, F1804), anti-FLAG M2 magnetic beads (Sigma, M8823), anti-Strep (Iba, 2-1507-001), anti-WDR11 (Thermo, PA5-25619), anti-snapin (Synaptic Systems, 148002), anti-muted (Aviva Systems Biology, OAAB00145), anti-pallidin (Proteintech, 10891–2-AP), goat anti-mouse IgG (H+L)-HRP conjugate (Bio-Rad, 172-1011), goat anti-rabbit IgG (H+L)-HRP conjugate (Bio-Rad, 170-8241), IRDye 680 goat anti-mouse IgG (H+L) (Li-Cor, 926-32220), and IRDye 800CW goat anti-rabbit IgG (H+L) (Li-Cor, 926-32211).
A rabbit polyclonal anti-dysbindin antibody was raised (Covance, NJ) against the C-terminal region of rat dysbindin (amino acids 202–350) and affinity-purified before use.
Mice
All of the experimental procedures were performed with C57BL/6 strain mice in accordance with the ILAR Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee of the National Institute of Mental Health.
Generation of BAC Transgenic Mice
The BAC clone (RP24-277I15) containing the mouse dysbindin gene was selected by BACFinder (http://www.mitocheck.org/cgi-bin/BACfinder)42 and obtained from BACPAC Resources Center (BPRC), CHORI (https://bacpac.chori.org/). The dysbindin gene in the BAC clone was manipulated by recombineering (recombination-mediated genetic engineering) according to the protocol developed by National Cancer Institute (NCI)-Frederick (http://ncifrederick.cancer.gov/research/brb/recombineeringInformation.aspx),43 including adding an N-terminal tag and a C-terminal reporter gene Venus (IRES-GB2-Venus). The N-terminal 3×FLAG-6×His-TEV-Strep tag (MDYKDHDGDYKDHDIDYKDDDDKHHHHHHENLYFQSWSHPQFEK) including the flanking homology arms for recombination was constructed by synthetic overlap PCR using the following DNA oligos: 5′-CCC GAC GGC GTC CGA GGG CGC GGT GGC GCG AGG CCT GAG GGA GGG GAC GCG ATG GAC TAC-3′, 5′-CAT CGC GTC CCC TCC CTC AGG CCT CGC GCC ACC GCG CCC TCG GAC GCC GTC GGG A-3′, 5′-AAA GAC CAT GAC GGT GAT TAT AAA GAT CAT GAC ATC GAC TAC AAG GAT GAC GAT GAC AAG-3′, 5′-ATC GTC ATC CTT GTA GTC GAT GTC ATG ATC TTT ATA ATC ACC GTC ATG GTC TTT GTA GTC-3′, 5′-CAT CAC CAT CAC CAT CAC GAG AAT CTT TAT TTC CAA TCC TGG AGC CAC CCC CAG TTC GAA-3′, 5′-CTG GGG GTG GCT CCA GGA TTG GAA ATA AAG ATT CTC GTG ATG GTG ATG GTG ATG CTT GTC-3′, 5′-AAA CTG GAG ACC CTG CGC GAG CGG CTG CTG AGC GTA CAG CAG GAT TTC ACC TCC A-3′, and 5′-GGA GGT GAA ATC CTG CTG TAC GCT CAG CAG CCG CTC GCG CAG GGT CTC CAG TTT TTC GAA-3′. The PCR product was cloned into pCR2.1 vector and verified by DNA sequencing. The linear DNA of the tag for recombineering was generated by PCR using the pCR2.1 construct as a template and the following primer pair: 5′-CCC GAC GGC GTC CGA GG-3′ and 5′-GGA GGT GAA ATC CTG CTG TAC GCT CAG C-3′. After recombineering, the insertion of the N-terminal tag was confirmed by DNA sequencing, and the C-terminal reporter gene Venus was added according to the same recombineering procedures. The GB2-Venus-kan was synthesized by overlap PCR using the following primer pairs: GB2-Venus-spacer (5′-AGC ACG TGT TGA CAA TTA ATC ATC GGC ATA GTA TAT CGG CAT AGT ATA ATA CGA CAA GGT GAG GAA CTA AAC CAA TGG TGA GCA AGG GCG AGG-3′ and 5′-GTG ATG GTG ATG ATG GTG GTG ATG CTT GTA CAG CTC GTC CAT GCC GAG AGT GAT CCC G-3′), spacer-neo (5′-CAT CAC CAC CAT CAT CAC CAT CAC GGA TCG GCC ATT GAA CAA GAT GGA TTG-3′ and 5′-AAT CAT GAT CAG AAG AAC TCG TCA AGA AGG CGA TAG A-3′), and GB2-Venus-kan with BspHI sites at both ends (5′-AAT CAT GAG CAC GTG TTG ACA ATT AAT CAT CGG CAT AG-3′ and 5′-AAT CAT GAT CAG AAG AAC TCG TCA AGA AGG CGA TAG A-3′). To generate IRES-GB2-Venus-kan, the EGFPcre-Frt-kan-Frt cassette after the IRES in the pIGCN21 construct (one of the recombineering reagents provided by NCI) was replaced by GB2-Venus-kan through NcoI and BspHI sites. The linear DNA of the C-terminal reporter Venus (IRES-GB2-Venus) for recombineering was generated by PCR using the IRES-GB2-Venus-kan construct as a template and the following primer pair containing the homologous targeting sequences: 5′- CAG GTG TCA GTG ATG ACA GTG ACC AGT GTG ACT CAA CTC AGG ACA TTT AAT CCG GTT ATT TTC CAC CAT ATT GCC GTC-3′ and 5′-TCT AAG CAC CTG GCT TTG CAG GAG GAT TCT GCG CAC AGC TGA CAA GTC ACT TAC TTG TAC AGC TCG TCC ATG CCG-3′. Finally, the insertion of IRES-GB2-Venus after the stop codon of the dysbindin gene was verified by DNA sequencing. The BAC construct was linearized and injected into zygotes from C57BL/6 mice. Microinjected zygotes were implanted into pseudopregnant C57BL/6 female mice to generate transgenic mice.
Sucrose Gradient Ultracentrifugation and Western Blot Analysis
Two mouse brains without cerebellums (postnatal day 14 or P14) were homogenized in buffer A (0.32 M sucrose, 1 mM NaHCO3, 1 mM MgCl2, 0.5 mM CaCl2, 5 mM NaF, and 1 mM Na3VO4; 4 mL/g of brain tissue) supplemented with protease inhibitors using a Dounce tissue grinder (tight pestle, 20 strokes). Cell debris and nuclei were removed by low-speed centrifugation at 1400g for 10 min. The crude membrane fraction (P2) in the supernatant was collected by centrifugation at 13 800g for 10 min. The pellet (P2) was washed twice with cold PBS. Half of the P2 fraction was subjected to chemical cross-linking with 1 mM (final concentration) DSP in PBS for 15 min on ice. The reaction was quenched by adding 1 M Tris-HCl, pH 7.5, to a final concentration of 100 mM and incubated for additional 15 min on ice. Membrane-bound proteins were solubilized in TBS supplemented with 1% Triton X-100 (1 mL/g of brain tissue) on ice for 15–30 min and clarified by centrifugation at 26 000g for 20 min. The resulting supernatant was collected and separated on a continuous sucrose gradient (10–40%) by centrifugation at 55 000 rpm (avg 286 794g) with a Beckman SW55Ti rotor at 4 °C for 5 h. Following centrifugation, 300 μL fractions were collected from top to bottom. Equal aliquots from individual fractions were resolved by SDS-PAGE and analyzed by immunoblotting. BSA (66 kDa, 4.4 S) and thyroglobulin (669 kDa, 19.4 S) were used as size standards for sucrose gradient ultracentrifugation.
Affinity Purification of Dysbindin Complexes
Forty whole brains without cerebellums from wild-type (negative control) or BAC transgenic mice (ages from P14 to P17) were used for affinity purification. The P2 fraction was cross-linked by DSP, and the membrane-bound proteins were extracted as described previously. The cleared extraction was incubated with monoclonal anti-FLAG-M2 magnetic beads (100 μL, Sigma) at 4 °C overnight on a rotator. Anti-FLAG beads were pelleted by a magnetic rack and washed five times with TBS containing 1% Triton X-100. The bound proteins were eluted with 3×FLAG peptide according to the manufacturer’s instruction (Sigma). The purified complexes were resolved on a SDS-PAGE gel. 10% of the complexes were used for immunoblot analysis, and 90% were used for SYPRO Ruby staining (Bio-Rad). Protein bands were visualized using a UV light, and the image was taken using an LAS-1000plus system (Fujifilm). The whole lane was excised into 21 fractions and stored at −80 °C for further mass spectrometry analysis.
Mass Spectrometry
The in-gel tryptic digestion was carried out by following the protocol described previously.44 Briefly, the gel bands were cut into 1 mm3 pieces, rinsed, and dehydrated, and the protein was reduced with DTT and alkylated with iodoacetamide in the dark prior to overnight digestion with trypsin at 37 °C in 50 mM ammonium bicarbonate. The concentrated peptides were analyzed on an LTQ Orbitrap Velos (Thermo Fisher Scientific, San Jose, CA) coupled with an Eksigent nanoLC-Ultra 1D plus system (Dublin, CA). Peptides were separated on a PicoFrit analytical column (100 mm long, ID 75 μm, tip i.d. 10 μm, packed with BetaBasic 5 μm 300 Å particles, New Objective, Woburn, MA) using a 35 min linear gradient of 5–35% ACN in 0.1% FA at a flow rate of 250 nL/min. Mass analysis was carried out in data-dependent analysis mode, where MS1 scanned the full MS mass range from m/z 300 to 2000, at 30 000 mass resolution, and 10 CID MS2 scans were sequentially carried out in the Orbitrap and the ion trap, respectively.
Database Searching
Tandem mass spectra were extracted, charge-state deconvoluted, and deisotoped by Extract_msn from Xcalibur version 2.0. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 1.3.0.339) and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1)). Mascot was set up to search Mascot5_Sprot_Mus musculus (house mouse) (12 551 entries) (only “Mudpit_A01”) assuming the digestion enzyme is trypsin, Mascot5_Sprot_Mus musculus (house mouse) (12 971 entries) (only “Mudpit_B01”) also assuming trypsin, and Mascot5_Sprot_Mus musculus (house mouse) (13 351 entries) (only “Mudpit_C01”) also assuming trypsin. X! Tandem was set up to search a subset of the uniprot_sprot database, also assuming trypsin. Mascot and X! Tandem were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 25 PPM. Carbamidomethyl of cysteine was specified in Mascot and X! Tandem as a fixed modification. Deamidated of asparagine and glutamine, oxidation of methionine, and acetylation of the N-terminus were specified in Mascot and X! Tandem as variable modifications.
Criteria for Protein Identification
Scaffold (version Scaffold_4.3.2, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm.45 Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.46 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The resulting peptide false discovery rate (FDR) and protein FDR were 0.0 and 0.1%, respectively, using a decoy database.
Plasmid Construction
The ORF cDNAs of dysbindin was tagged with 3×FLAG at the N-terminus and amplified from the cDNA clone IMAGE 4139934 (ATCC) using the primer pair 5′-TTT GGA TCC GCC GCC ACC ATG GAC TAC AAA GAC CAT GAC GGT GAT TAT AAA GAT CAT GAC ATC GAC TAC AAG GAT GAC GAT GAC AAG GGC GGT GGC GGT ATG CTG GAG ACC CTT CGC GAG-3′ and 5′-TTT TCT AGA TTA AGA GTC GCT GTC CTC ACC ACC-3′ and was cloned into pEF-ENTR B-term vector between the BamHI and XbaI sites.47 The ORF cDNA of WDR11 was amplified from the cDNA clone IMAGE 30346203 (Life Technologies) using the primer pair 5′-TTG GAT CCG CCA CCA TGT TGC CCT ACA CAG TGA ACT TCA AGG-3′ and 5′-TTG CGG CCG CCC TCT TCA ATG GGT TCT TCC TTG GGG G-3′ and was cloned into pEF-ENTR B-term vector (containing a V5 tag at the C-terminus) between the BamHI and NotI sites. The ORF cDNA of FAM91A1 was amplified from the cDNA clone IMAGE 9092514 (ATCC) using the primer pair 5′-TTG GTA CCG CCA CCA TGA ACA TCG ACG TTG AGT TCC ACA TCC-3′ and 5′-TTG CGG CCG CCC TGC AAA TGG AGA CCA GCA ATG AGC-3′ and was cloned into pEF-ENTR B-term vector between the KpnI and NotI sites. The ORF cDNA of PSMD9 was amplified from the cDNA clone IMAGE 2961610 (ATCC) using the primer pair 5′-TTG GAT CCG CCA CCA TGT CCG ACG AGG AAG CGA GG-3′ and TTG CGG CCG CCT CTT TGC AGA GGA ATA ATG TTG CAG CCC-3′ and was cloned into pEF-ENTR B-term vector between the BamHI and NotI sites. The cDNA clone of PSMA4 (IMAGE 3592024) was obtained from ATCC, and a Strep tag was introduced by PCR using the primer pair 5′-GGT GGC TGG AGC CAC CCC CAG TTC GAA AAG TAG ACA GAA TCA CGG ATT TTA TAA CTC CTT-3′ and 5′-/5Phos/TTT ATC CTT TTC TCT CTG TTC TTT TTC TTT CTT C-3′, followed by self-ligation. All constructs were verified by DNA sequencing.
Cell Transfection and Co-immunoprecipitation Analysis
HEK293T cells were grown in Dulbecco’s modified Eagle’s medium with 10% FBS. Cells were transfected using GenJet in vitro DNA transfection reagent (Ver. II) according to the manufacturer’s instructions (SignaGen Laboratories). For coimmunoprecipitation analysis, transfected HEK293T cells were washed with PBS, harvested, and lysed with a buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10 mm NaF, 10 mm Na3VO4, 1% Triton X-100, and protease inhibitor mixture (Roche Applied Science). Lysates were spun at 16 100g for 20 min at 4 °C, and the supernatant was incubated with anti-FLAG M2 magnetic beads (Sigma) for 2 h at 4 °C on a rotator. The resin was then washed six times with the same buffer, and the bound proteins were eluted with 3×FLAG peptide. The eluates were then subjected to SDS-PAGE and immunoblot analysis.
Proteasome Activity Assay
The proteasome activity assay for tissue samples was performed as described,48 with modifications, using the Proteasome-Glo Assay System with the substrate for chymotrypsin-like activity (Promega). The soluble cytoplasm and P2 fractions of wild-type and sdy mouse brains (P14–15) were extracted as described previously without cross-linking. Membrane-bound proteins in the P2 fraction were solubilized in TBS supplemented with 1% Triton X-100 (1 mL/g of brain tissue) on ice for 15–30 min and clarified by centrifugation. The extracted proteins were quantified and diluted to a concentration of 0.2 μg/μL with PBS containing 5 mM EDTA. Fifty microliters (10 μg) of the diluted sample was used for each assay with equal volume of Proteasome-Glo substrate mix (50 μL) in a 96-well plate. After mixing, the plate was incubated at room temperature for 60 min, and luminescence generated as a result of the cleavage of the substrate was measured with a LumiCount microplate luminometer (Packard BioScience, USA). The background non-specific activity was measured in the same way but with the addition of 30 μM of the irreversible and specific proteasome inhibitor adamantane-acetyl-(6-aminohexanoyl) 3-(leucinyl)3-vinyl-(methyl)-sulfone (AdaAhx3L3VS, Calbiochem). The specific proteasomal activity was calculated as follows: specific proteasomal activity = total peptidase activity (no inhibitor) – non-specific peptidase activity (with inhibitor). All assays were performed in triplicate. Student’s two-tailed t test was used for statistical analysis (p < 0.05 considered significant).
Results
Dysbindin Has Three Isoforms Associating with Different Protein Complexes in the P2 Synaptosome Fraction of Mouse Brain
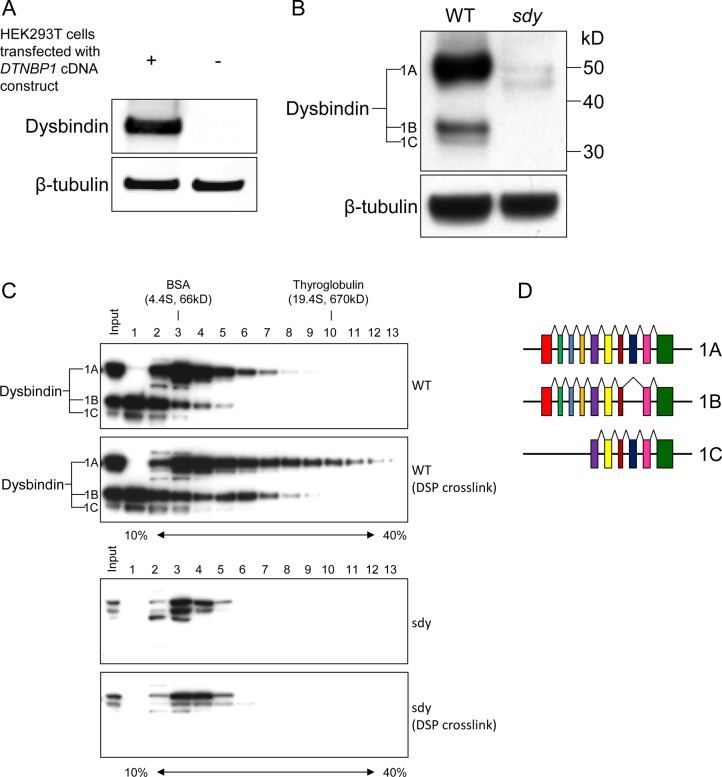
To investigate the interactome of dysbindin in the synaptosome, we first characterized dysbindin-containing protein complexes in the P2 synaptosome fraction. To this end, a polyclonal antibody against the C-terminal region (amino acid 202–350) of dysbindin was generated in rabbits and affinity purified. As shown in Figure 1A, this antibody specifically reacts with the recombinant dysbindin expressed in HEK293T cells. When it was tested on the mouse brain tissue extract, three specific dysbindin isoforms were detected with the sizes of about 50, 35, and 32 kDa (Figure 1B). We found that this antibody also detects non-specific bands around 40–50 kDa in the sample from sandy mouse, but they are much weaker than the specific bands (Figure 1B,C). In human, there are three major isoforms, dysbindin-1A, -1B, and -1C, that can be detected by immunoblotting.24 In the UniProt database, three mouse dysbindin isoforms were annotated (Figure 1D), including 1A (352 amino acids or aa) and 1C (271 aa) that are orthologues of human dysbindin isoforms,24 as well as a unique isoform (300 aa) that has not been named. According to their sizes, it is likely that the polyclonal antibody detects all these isoforms. Therefore, we designated these isoforms as dysbindin-1A, -1B, and -1C, from large to small size, respectively. Using this antibody as a detection tool, we profiled dysbindin-associated complexes in P2. The insoluble P2 fraction of mouse brain was isolated, cross-linked by dithiobis(succinimidyl proprionate) (DSP, omitted for control), and then solubilized using 1% Triton X-100 in TBS. DSP cross-linking can stabilize transient and/or weak protein–protein interactions under harsh isolation conditions (i.e., with detergent) and has been applied successfully to isolate labile multiprotein complexes.49,50 The cleared P2 lysate was separated by ultracentrifugation on a 10–40% linear sucrose gradient (SG). The sucrose gradients were fractionated into 14 equal fractions from top to bottom, and an equal aliquot of each fraction was subjected to immunoblot analysis using the anti-dysbindin antibody. As shown in Figure 1C, dysbindin-1A was concentrated close to BSA (66 kDa, 4.4S) without DSP cross-linking. The overall profile shifted to higher density fractions with DSP cross-linking, indicating that dysbindin-1A is present in large protein complex(es) that are labile to Triton X-100 extraction and DSP stabilizes these associations. There is no obvious peak in the shifted fractions after fractions 3 and 4, suggesting that dysbindin-1A may be present in multiple complexes that overlap in the sucrose gradient. Similarly, dysbindin-1B and -1C also shifted to higher density fractions, and a distinct complex peak of dysbindin-1B in fractions 5 and 6 can be observed after cross-linking (Figure 1C). Taken together, the SG analyses and DSP cross-linking demonstrate that dysbindin is present in macromolecular complexes in the P2 synaptosome fraction of brain. Consistent with previous study that dysbindin isoforms have distinct subsynaptic localization,23 our data suggest that dysbindin-1A, -1B, and -1C are present in different complexes. Dysbindin-1A is present in complex(es) larger than that of BLOC-1, which was estimated to be ∼230 kDa (5.2S).34,51
Figure 1.
Characterization of anti-dysbindin antibody, and sucrose density gradient ultracentrifugation (SG) analysis of dysbindin-containing complex(es) in the P2 synaptosome fraction of mouse brain. (A) Immunoblot analysis of extract from HEK293T cells transfected with DTNBP1 expression construct using the custom-made polyclonal anti-dysbindin antibody. The extract from untransfected HEK293T cells was used as a negative control. (B) Immunoblot analysis of total extracts from wild-type (WT) and sandy (sdy) mouse brains using the same antibody as that in panel A. (C) SG analysis of dysbindin-containing complex(es) in the P2 synaptosome fraction of wild-type mouse brain with and without DSP cross-linking. Equal aliquots from individual fractions were resolved by SDS-PAGE and analyzed by immunoblotting using the anti-dysbindin antibody. β-Tubulin served as a loading control in panels A and B. Samples from sandy mice served as a negative control to show the specificity of immunoblot analysis in panels B and C. (D) Structures of the alternative splicing transcripts based on the annotated mouse dysbindin isoforms in the UniProt database. The colored boxes indicate the alternative splicing coding exons. Sizes of the structures are not scaled precisely.
Generation of BAC Transgenic Mice That Express Tagged Dysbindin
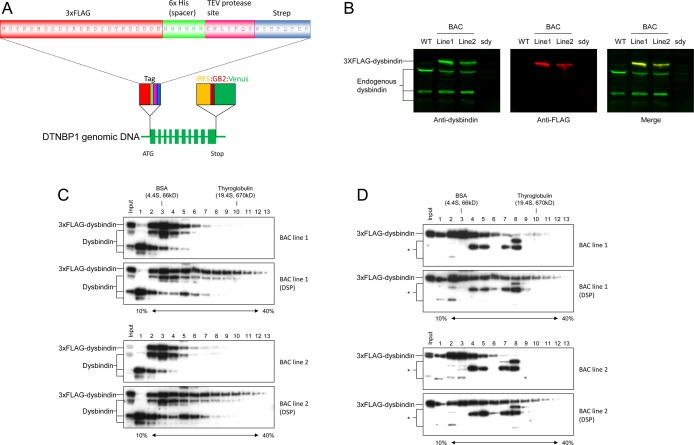
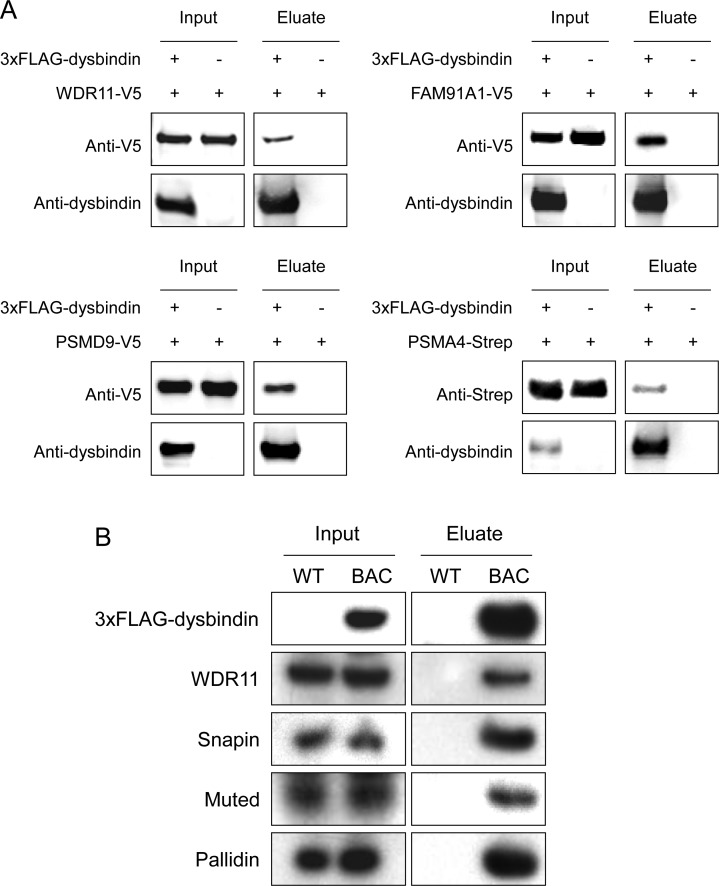
The finding that dysbindin is associated with large complexes in P2 prompted us to further explore the components in the complexes. Although more than 100 dysbindin-binding partners have been described in the literature,12 there are no reported systematic proteomics analysis of in vivo (endogenous) dysbindin complexes in the brain to date. The lack of reports on this topic is due likely to the absence of suitable antibodies available for the immunopurification of dysbindin from brain extracts, prohibiting isolation and characterization of the dysbindin complex for proteomics analyses. We tested our anti-dysbindin antibody and found that it was also not suitable for immunopurification (data not shown). To overcome this issue, we generated bacterial artificial chromosome (BAC) transgenic mice in which a tagged dysbindin is expressed through its genomic DNA and under the control of its native regulatory sequences.52,53 The BAC clone possessing the dysbindin genomic DNA was epitope-tagged at the N-terminus by recombineering in Escherichia coli (Figure 2A),43 and BAC transgenic mice were made by the pronuclear microinjection method.53 To this end, two BAC lines with stable expression of tagged dysbindin were obtained. The BAC transgenic mice appear normal and express both tagged and wild-type dysbindin (Figure 2B). However, only tagged dysbindin-1A could be detected because either the 1B and 1C isoforms do not contain the N-terminal end of isoform 1A after splicing or normal splicing was disrupted by the inserted tag or reporter gene. On the basis of Figure 2B, the expression levels of the tagged dysbindin-1A are about 2.3- and 1.7-fold of that of the untagged dysbindin-1A in lines 1 and 2, respectively. As multiple BACs are commonly inserted into the mouse genome after microinjection,54 it is likely that both BAC lines possess multiple BAC transgenes (the actual BAC copy numbers were not determined in this study). In the SG analysis, the tagged dysbindin is cofractionated with wild-type dysbindin (Figure 2C using anti-dysbindin antibody and 2D using anti-FLAG antibody), indicating that the tagged dysbindin protein is functionally incorporated into the endogenous dysbindin complexes. Therefore, the BAC transgenic lines provide us with an optimal source for affinity purification of the endogenous dysbindin-associated complexes.
Figure 2.
Tagging cassettes for BAC recombineering and characterization of tagged dysbindin in the brains of BAC transgenic mouse lines. (A) The tag was inserted immediately after the ATG start codon and consists of a 3×FLAG tag for immunopurification and detection, a 6×His spacer for TEV protease cleavage, a tobacco etch virus (TEV) protease cleavage site, and a Strep tag for the second affinity purification and detection. The reporter cassette was inserted immediately after the stop codon and consists of an internal ribosome entry site (IRES) and a bacterial promoter (GB2) in front of the gene encoding Venus fluorescent protein. (B) Immunoblot analysis of total extracts from wild-type, sandy, and BAC transgenic mouse brains using anti-dysbindin and anti-FLAG antibodies. (C) SG analysis of dysbindin-containing complex(es) in the P2 synaptosome fraction of the BAC transgenic mouse brains with and without DSP cross-linking. Equal aliquots from individual fractions were resolved by SDS-PAGE and analyzed by immunoblotting using the anti-dysbindin antibody. (D) The blots in panel C were reprobed with the anti-FLAG antibody. * indicates nonspecific bands.
Proteomics Analysis of Membrane-Bound Dysbindin Complexes in P2
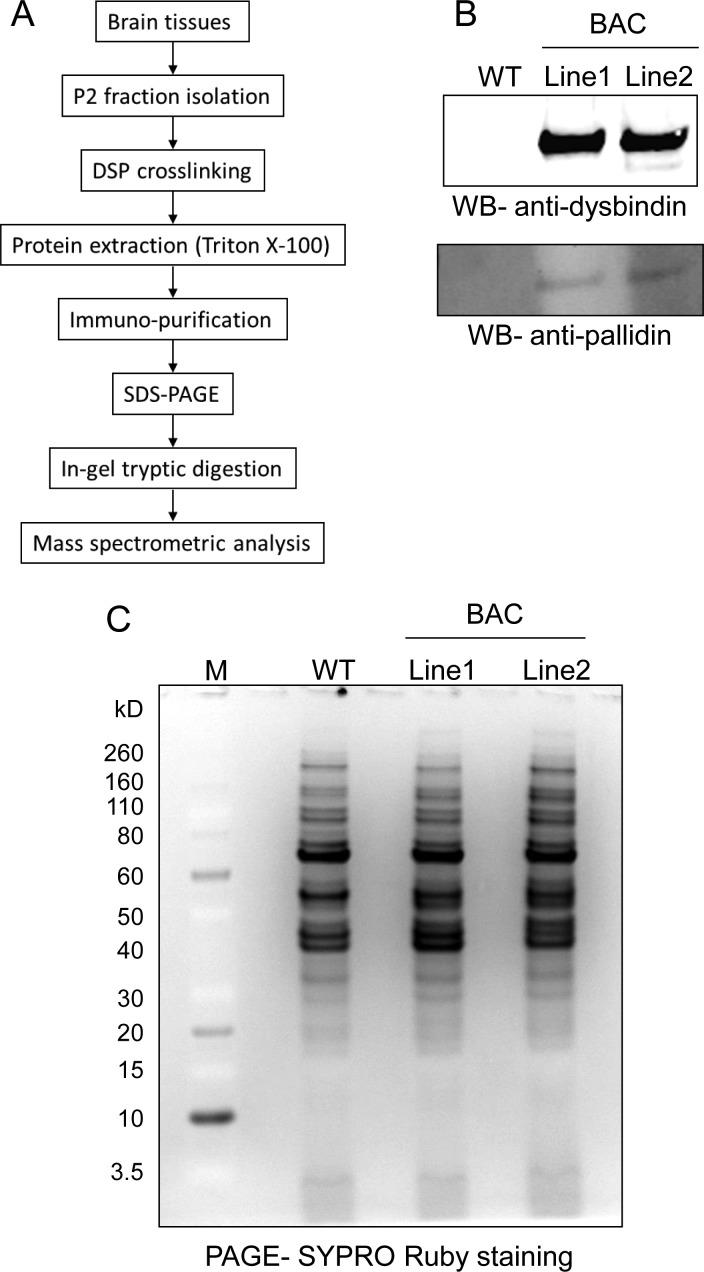
To identify the proteins associated with dysbindin, the dysbindin-containing complexes were immunopurified from the P2 fraction and analyzed by mass spectrometry. The proteomics analysis is shown schematically in Figure 3A. Because (1) dysbindin is not an abundant protein, (2) only a small fraction (∼10%) of dysbindin in the brain is present in the insoluble fraction,32 and (3) dysbindin is likely present in multiple complexes in the extraction (Figures 1C and 2C), we attempted to collect as much brain tissue as possible for purification in order to obtain sufficient material for mass spectrometric analysis. To this end, 40 brains (P14–P17) from each of the BAC lines and wild-type mice were collected for P2 isolation. The developmental stage at P14–P17 was selected because the onset of schizophrenia symptoms typically occurs in young adulthood and the expression of dysbindin is greatly reduced in adulthood.32 The isolated P2 fraction was subjected to DSP cross-linking followed by Triton X-100 extraction. The cleared lysate was then subjected to anti-FLAG purification. The bound complexes were eluted specifically with 3×FLAG peptides under native conditions, followed by SDS-PAGE analysis. Approximately 10% of the eluate was analyzed by immunoblotting with anti-dysbindin antibody and anti-pallidin (reprobing). Both tagged dysbindin and pallidin were specifically detected in the eluate of the BAC lines but not in that of the wild-type negative control mice, indicating that the affinity purifications were successful (Figure 3B). No wild-type dysbindin was detected in the purified complexes, suggesting that there is only one dysbindin in each complex stoichiometrically and that dysbindins do not exist in vivo as homomultimers.
Figure 3.
Immunopurification of dysbindin-associated protein complex(es) in the P2 fraction of brain. (A) Flowchart of purification procedures. (B) Immunoblot analysis of dysbindin-containing complex(es) purified from the P2 fraction of BAC transgenic mouse brains using anti-dysbindin and anti-pallidin antibodies. (C) SYPRO Ruby-stained SDS-PAGE gel of the purified dysbindin-containing complex(es). Wild-type served as a negative control in panels B and C.
For mass spectrometric analysis, 90% of the eluate was separated by SDS-PAGE, stained with SYPRO Ruby, and visualized using a LAS-1000plus system (Fujifilm) (Figure 3C). The entire protein lane from each sample in the SDS-PAGE was excised and analyzed by LC–MS/MS. The MS spectra containing the information on peptide masses and sequences were searched using the Mascot search engine against the mouse nonredundant protein database. The putative peptide results from two replicates were then consolidated and filtered using the Proteome Software Scaffold. Using the criteria of ≥95% probability for both protein and peptide and ≥2 peptides matching, 48 nonredundant proteins, including the bait dysbindin, were identified in both BAC lines but not in the wild-type control (for higher stringency, proteins with ≥1 peptide matching in the control were considered as background) (Table 1). Notably, all known members of the BLOC-1 complex (dysbindin, pallidin, muted, cappuccino, snapin, Bloc1s1, Bloc1s2, and Bloc1s3) were identified and ranked by Scaffold among the top 10 proteins in the list, suggesting that the BLOC-1 complex is the core or the most abundant complex of dysbindin in brain synaptosome. To our knowledge, this is the first time that all members of the BLOC-1 complex have been identified along with dysbindin by a single proteomics analysis of brain tissues. Only pallidin and snapin have been previously confirmed to be associated with dysbindin in the brain using coIP and immunoblot analysis.30,55
Table 1. Mass Spectrometric Identification of Proteins That Co-purify with Dysbindina.
| BAC line
1 |
BAC line 2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| no. | identified proteins | symbol | accession no. | molecular weight (kDa) | protein identification probability (%) | unique peptide | percent coverage (%) | protein identification probability (%) | unique peptide | percent coverage (%) |
| 1 | SNARE-associated protein Snapin | Bloc1s7 | SNAPN_MOUSE | 15 | 100 | 16 | 93.40 | 100 | 9 | 59.60 |
| 2 | Protein Muted | Bloc1s5 | MUTED_MOUSE | 21 | 100 | 24 | 85.90 | 100 | 17 | 83.20 |
| 3 | Dysbindin | Dtnbp1 | DTBP1_MOUSE | 40 | 100 | 13 | 40.30 | 100 | 13 | 40.30 |
| 4 | Protein cappuccino | Bloc1s4 | CNO_MOUSE | 23 | 100 | 16 | 85.60 | 100 | 15 | 65.60 |
| 5 | WD repeat-containing protein 11 | Wdr11 | WDR11_MOUSE | 136 | 100 | 44 | 48.50 | 100 | 27 | 30.50 |
| 6 | Biogenesis of lysosome-related organelles complex 1 subunit 1 | Bloc1s1 | BL1S1_MOUSE | 14 | 100 | 7 | 78.40 | 100 | 6 | 68.80 |
| 7 | Biogenesis of lysosome-related organelles complex 1 subunit 2 | Bloc1s2 | BL1S2_MOUSE | 16 | 100 | 14 | 83.20 | 100 | 13 | 79.70 |
| 8 | Biogenesis of lysosome-related organelles complex 1 subunit 3 | Bloc1s3 | BL1S3_MOUSE | 20 | 100 | 12 | 67.70 | 100 | 13 | 67.70 |
| 9 | Protein FAM91A1 | Fam91a1 | F91A1_MOUSE | 93 | 100 | 25 | 32.60 | 100 | 15 | 23.90 |
| 10 | Pallidin | Bloc1s6 | PLDN_MOUSE | 20 | 100 | 6 | 62.80 | 100 | 5 | 56.40 |
| 11 | AP-3 complex subunit beta-1 | Ap3b1 | AP3B1_MOUSE | 123 | 100 | 21 | 19.00 | 100 | 17 | 15.30 |
| 12 | Exocyst complex component 3 | Exoc3 | EXOC3_MOUSE | 86 | 100 | 15 | 19.30 | 100 | 3 | 4.64 |
| 13 | Vinculin | Vcl | VINC_MOUSE | 117 | 100 | 2 | 2.16 | 100 | 21 | 28.80 |
| 14 | Exocyst complex component 4 | Exoc4 | EXOC4_MOUSE | 111 | 100 | 14 | 18.60 | 100 | 7 | 10.80 |
| 15 | Proteasome subunit alpha type-4 | Psma4 | PSA4_MOUSE | 29 | 100 | 4 | 23.40 | 100 | 5 | 32.60 |
| 16 | Transgelin-2 | Tagln2 | TAGL2_MOUSE | 22 | 100 | 3 | 20.10 | 100 | 9 | 60.80 |
| 17 | Calcineurin subunit B type 1 | Ppp3r1 | CANB1_MOUSE | 19 | 100 | 6 | 62.90 | 100 | 4 | 29.40 |
| 18 | ATP-dependent RNA helicase DDX1 | Ddx1 | DDX1_MOUSE | 83 | 100 | 6 | 8.78 | 100 | 2 | 2.30 |
| 19 | Calnexin | Canx | CALX_MOUSE | 67 | 100 | 2 | 4.40 | 100 | 7 | 14.60 |
| 20 | E3 ubiquitin-protein ligase HUWE1 | Huwe1 | HUWE1_MOUSE | 483 | 100 | 2 | 0.62 | 100 | 3 | 0.98 |
| 21 | Vacuolar protein sorting-associated protein 16 homologue | Vps16 | VPS16_MOUSE | 95 | 100 | 8 | 11.60 | 100 | 2 | 3.34 |
| 22 | Syntaxin-1B | Stx1b | STX1B_MOUSE | 33 | 100 | 2 | 10.10 | 100 | 4 | 17.40 |
| 23 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase | Cdipt | CDIPT_MOUSE | 24 | 100 | 2 | 10.30 | 100 | 5 | 27.20 |
| 24 | TGF-beta-activated kinase 1 and MAP3K7-binding protein 3 | Tab3 | TAB3_MOUSE | 79 | 100 | 6 | 13.00 | 100 | 4 | 8.52 |
| 25 | Carbonic anhydrase 2 | Ca2 | CAH2_MOUSE | 29 | 100 | 4 | 23.50 | 100 | 3 | 13.50 |
| 26 | Rabphilin-3A OS=Mus musculus GN=Rph3a | Rph3a | RP3A_MOUSE | 75 | 100 | 3 | 8.37 | 100 | 4 | 9.40 |
| 27 | Peripheral plasma membrane protein CASK | Cask | CSKP_MOUSE | 105 | 100 | 3 | 3.13 | 100 | 3 | 3.24 |
| 28 | Serine/threonine-protein kinase MARK1 | Mark1 | MARK1_MOUSE | 88 | 100 | 4 | 7.17 | 100 | 5 | 8.81 |
| 29 | Dedicator of cytokinesis protein 3 | Dock3 | DOCK3_MOUSE | 233 | 100 | 2 | 1.43 | 100 | 3 | 1.78 |
| 30 | Neurobeachin | Nbea | NBEA_MOUSE | 327 | 100 | 2 | 1.26 | 100 | 5 | 2.08 |
| 31 | Myelin proteolipid protein | Plp1 | MYPR_MOUSE | 30 | 100 | 2 | 6.86 | 100 | 2 | 8.66 |
| 32 | Serine/threonine-protein kinase MARK2 | Mark2 | MARK2_MOUSE | 86 | 100 | 4 | 7.35 | 100 | 2 | 3.61 |
| 33 | SEC14 domain and spectrin repeat-containing protein 1 | Sestd1 | SESD1_MOUSE | 79 | 100 | 2 | 2.73 | 100 | 5 | 7.18 |
| 34 | 26S proteasome non-ATPase regulatory subunit 8 | Psmd8 | PSMD8_MOUSE | 40 | 100 | 2 | 6.80 | 100 | 2 | 6.52 |
| 35 | V-type proton ATPase subunit D | Atp6v1d | VATD_MOUSE | 28 | 100 | 3 | 19.00 | 100 | 3 | 19.00 |
| 36 | ATP synthase subunit f, mitochondrial | Atp5j2 | ATPK_MOUSE | 10 | 100 | 2 | 23.90 | 100 | 2 | 23.90 |
| 37 | 26S proteasome non-ATPase regulatory subunit 9 | Psmd9 | PSMD9_MOUSE | 25 | 100 | 3 | 13.10 | 100 | 3 | 13.10 |
| 38 | Beta-adrenergic receptor kinase 1 | Adrbk1 | ARBK1_MOUSE | 80 | 100 | 2 | 2.76 | 100 | 4 | 6.68 |
| 39 | C-Jun-amino-terminal kinase-interacting protein 3 | Mapk8ip3 | JIP3_MOUSE | 148 | 100 | 2 | 1.50 | 100 | 3 | 2.24 |
| 40 | Uncharacterized protein C6orf203 homologue | C6orf203 | CF203_MOUSE | 28 | 100 | 2 | 9.17 | 100 | 2 | 9.17 |
| 41 | Hyccin | Fam126a | HYCCI_MOUSE | 57 | 100 | 3 | 7.10 | 100 | 2 | 4.80 |
| 42 | Dynamin-3 | Dnm3 | DYN3_MOUSE | 97 | 100 | 8 | 9.62 | 100 | 8 | 10.80 |
| 43 | 1-Acyl-sn-glycerol-3-phosphate acyltransferase gamma | Agpat3 | PLCC_MOUSE | 43 | 100 | 2 | 5.05 | 100 | 3 | 6.65 |
| 44 | 39S ribosomal protein L41, mitochondrial | Mrpl41 | RM41_MOUSE | 15 | 100 | 2 | 20.00 | 100 | 2 | 20.70 |
| 45 | Protein fat-free homologue | Vps51 | FFR_MOUSE | 86 | 100 | 3 | 4.09 | 100 | 2 | 3.20 |
| 46 | Elongation factor 1-delta | Eef1d | EF1D_MOUSE | 31 | 100 | 2 | 9.25 | 100 | 2 | 9.25 |
| 47 | Catenin delta-2 | Ctnnd2 | CTND2_MOUSE | 135 | 100 | 2 | 1.52 | 100 | 2 | 1.84 |
| 48 | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform | Ppp2cb | PP2AB_MOUSE | 36 | 100 | 12 | 47.90 | 100 | 11 | 38.50 |
Only the proteins identified in both BAC transgenic lines but not in wild-type control were selected.
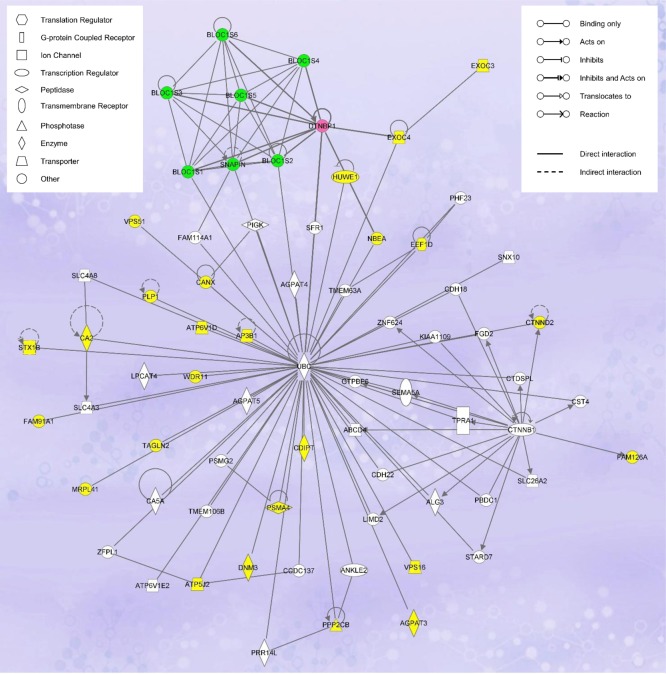
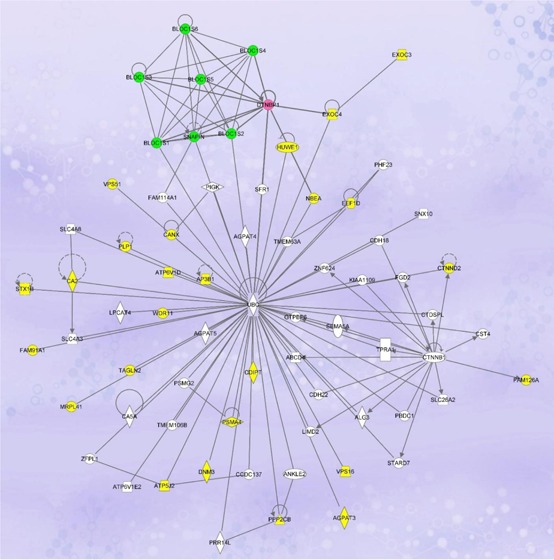
On the basis of the information retrieved from UniProt, NCBI, and/or Gene Ontology databases, we grouped the putative dysbindin-associated proteins into seven categories with regard to their functions: trafficking/transport (23 proteins), signaling (11 proteins), ubiquitin-proteasome system (4 proteins), transcription regulation (4 proteins), cell adhesion (3 proteins), lipid metabolism (2 proteins), and others with unique or unknown functions (6 proteins) (Supporting Information, Table S1). Pathway analysis was used to interrogate the functional networks associated with the putative 47 dysbindin-associated proteins (Ingenuity Pathway Analysis software, IPA, Ingenuity Systems, www.ingenuity.com). The network with the highest association score (score 78, focus molecule 33) revealed by the IPA is that devoted to “Dermatological Diseases and Conditions, Developmental Disorder, Hereditary Disorder” (Figure 4). The central player of this network is ubiquitin. Twenty seven of the identified proteins, including dysbindin and three other components of BLOC-1, have direct connections with ubiquitin. The other network involved is “Cell-Mediated Immune Response, Cellular Development, Cellular Function and Maintenance” (score 28, focus molecule 15) (Supporting Information, Figure S1). Key molecules in this network include Akt, ERK, P38 MAPK, Jnk, ADRBK1, Pkc, VCL, 26S proteasome, and others. Interestingly, both networks suggest that dysbindin is functionally interrelated to the ubiquitin-proteasome system (UPS).
Figure 4.
Ingenuity Pathway Analysis of the putative dysbindin-associated proteins showing the highest scoring networks (Dermatological Diseases and Conditions, Developmental Disorder, Hereditary Disorder) of the 48 proteins identified in this study. Colored nodes indicate the identified proteins used in the pathway analysis. Dysbindin is highlighted in pink, and the other BLOC-1 components, in green.
Validation of Mass Spectrometry Results
To verify the result from mass spectrometry analysis, we confirmed the interactions of dysbindin with several of the identified proteins using coIP followed by immunoblot analysis. There are two common approaches used to validate protein interaction: (1) coIP of two epitope-tagged proteins overexpressed in cultured cells and (2) coIP from brain tissue extract directly. The second approach tests endogenous interactions but is limited by the availability of working antibodies. For the first approach, we selected several novel dysbindin-associated candidates identified by our proteomics analysis, including WDR11 (WD repeat domain 11), FAM91A1 (family with sequence similarity 91, member A1), PSMD9 (proteasome 26S subunit, non-ATPase, 9), and PSMA4 (proteasome subunit, alpha type, 4). WDR11 and FAM91A1 are the two other non-BLOC-1 subunits of the top 10 dysbindin-associated proteins (Table 1). We also tested the association of dysbindin with the 26S proteasome subunits, as three of them (PSMA4, PSMD8, and PSMD9) were identified by our proteomics study. PSMA4 was selected because it is ranked highest among the three proteasome subunits, and PSMD9 was selected because several genetic studies suggest its linkage to depression and schizophrenia.56−58 To this end, we cotransfected HEK293T cells with the following pairs of expression vectors: 3×FLAG-dysbindin/WDR11-V5, 3×FLAG-dysbindin/FAM91A1-V5, 3×FLAG-dysbindin/PSMD9-V5, and 3×FLAG-dysbindin/PSMA4-Strep, followed by coIP with anti-FLAG M2 affinity resin and immunoblot analyses with anti-V5 (or anti-Strep for PSMA4) and anti-dysbindin antibodies. Only the constructs expressing candidate dysbindin-interacting proteins were transfected in the negative control cells. Using this approach, we confirmed the associations of WDR11, FAM91A1, PSMA4, and PSMD9 with dysbindin (Figure 5A). Interestingly, dysbindin was significantly reduced when cotransfected with PSMA4 (Figure 5A). As dysbindin can be ubiquitinated by the E3 ubiquitin ligase TRIM32,59 overexpression of PSMA4 could increase proteasome activity and degradation of dysbindin. We further tested the endogenous interaction of dysbindin with several candidates, for which working antibodies are available, using the second approach. To this end, the brain P2 extracts of the BAC transgenic and wild-type mice were subjected to coIP with anti-FLAG M2 affinity resin followed by immunoblot analyses with antibodies against the selected proteins. As shown in Figure 5B, the second approach confirmed the associations of WDR11, snapin, muted, and pallidin with dysbindin.
Figure 5.
Conformation of mass spectrometry-identified proteins by coIP and immunoblot analysis. (A) HEK293T cells were cotransfected with different combinations of expression vectors as indicated, followed by coIP using anti-FLAG beads and immunoblot analyses with anti-dysbindin, anti-V5, or anti-Strep antibodies. (B) The protein extracts from the P2 fractions of wild-type and BAC transgenic mouse brains were immunoprecipitated with anti-FLAG beads. The purified complexes were subjected to immunoblot analyses using antibodies against dysbindin, WDR11, snapin, muted, and pallidin. Wild-type was used as a negative control. Equal aliquots of the protein extracts were loaded in each analysis in panels A and B.
Proteasomal Activity Is Reduced in the P2 Fraction of the Brain of Sandy Mice
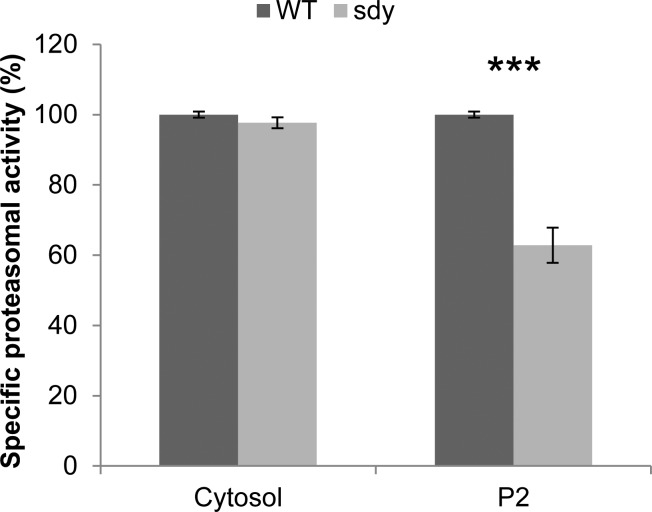
Protein degradation via UPS is a universal mechanism in maintaining protein homeostasis in eukaryotic cells. In addition to de novo protein synthesis, growing evidence suggests that UPS also plays an essential role in the regulation of synaptic connectivity and plasticity.60−62 The association of dysbindin with proteasome subunits suggests that it may have functional linkages to UPS. To test this hypothesis, we investigated whether proteasomal activity is affected in the brain of sandy mice. As proteasomes are localized to both the cytosol and synaptosome in the brain,63 we measured the proteasomal activities in the protein extracts from both fractions. Interestingly, we found that the specific proteasomal activity (see Materials and Methods for specific proteasomal activity measurement) is significantly decreased to ∼60% in the P2 fraction of sandy mice compared with wild-type, whereas the activity is unchanged in the cytosol (Figure 6). Taken together, our data suggest that dysbindin may regulate proteasomal activity in synapses by interacting with proteasome subunits.
Figure 6.
Specific chymotrypsin-like proteasomal activities in the extracts of soluble and insoluble fractions of brains of wild-type and sandy mice. The proteasome activity assay was performed using the Proteasome-Glo Assay System with the substrate for chymotrypsin-like activity (Promega). The soluble and insoluble fractions of wild-type and sdy mouse brains (P14–15) were extracted, quantified, and diluted in assay buffer with equal concentration. Ten micrograms of each sample was used and incubated with Proteasome-Glo substrate mix with or without specific proteasome inhibitor (AdaAhx3L3VS) in a 96-well plate for 60 min at RT, followed by luminescence measurement with a plate reader. The specific proteasomal activity was calculated as follows: specific proteasomal activity = total peptidase activity (no inhibitor) – non-specific peptidase activity (with inhibitor). The activities in the soluble or insoluble fraction are given as percentages of the mean activity of the WT. Student’s two-tailed t test was used for statistical analysis. n = 3; assay performed in triplicate; ***p < 0.001, compared with wild-type.
Discussion
Accumulating evidence obtained from the studies of putative SCZ risk genes, such as dysbindin, have revealed their common functions in synaptic transmission and neuroplasticity.11 In the present study, we targeted investigation of the dysbindin-associated proteome in the P2 synaptosome fraction of mouse brains. Because the synaptosome contains the molecular machinery essential for synaptic transmission, elucidating the associated proteome of dysbindin in the synaptosome should provide insights into the role of dysbindin in synaptic functions, which in turn may help to understand how changes in the expression of dysbindin contribute to the impaired synaptic connectivity typically observed in SCZ patients. It should be noted that the synaptosome is highly enriched in the P2 fraction.41 However, it is also known to contain mitochondria and light membranes. Despite extensive washing during the P2 preparation, we cannot eliminate the presence of minimal cytosolic protein contamination.
Dysbindin is a soluble protein, and its localization in the insoluble synaptosome suggests that it may be targeted to the membrane structures through protein–protein interactions with other proteins on the membrane. Therefore, it is likely that dysbindin exists as a component of protein complexes in the synaptosome. As the characterization of dysbindin-containing complex in the insoluble synaptosome has not yet been reported, we generated a polyclonal antibody against dysbindin and profiled the dysbindin complexes in the P2 synaptosome fraction using chemical cross-linking and sucrose density gradient ultracentrifugation. Overall, these data suggest that (1) there are 3 dysbindin isoforms in mouse brain migrated at about 50, 35, and 32 kDa by SDS-PAGE, (2) the 3 isoforms are present in different complexes, (3) dysbindin complexes are labile to Triton X-100 extraction, (4) DSP is useful to stabilize dysbindin complexes in P2, and (5) part of dysbindin is present in complexes with sizes larger than that of BLOC-1, indicating that dysbindin in P2 does not simply exist as a stable component of BLOC-1 as studies of soluble cytosol had suggested.32
In human, there are three reported dysbindin isoforms,24 but only two isoforms from mouse could be detected previously, with the sizes by SDS-PAGE of about 50 and 40 kDa, as well as 50 and 33 kDa in two independent studies.23,26 It is not clear whether the discrepancy in the sizes is due to the variations of electrophoresis and protein markers or if the 40 and 33 kDa proteins are actually different isoforms. The size of the largest isoform (dysbindin-1A) is consistent with previous studies despite the theoretical molecular weight of the dysbindin-1A being about 40 kDa.12,23,26 As of the time that this article was prepared, five human dysbindin isoforms, dysbindin-1A, -1B, -1C, -1D, and -1E (NP_115498, NP_898861, NP_001258596, NP_001258597, and NP_001258598, respectively), are listed in NCBI Entrez Protein database and three in UniProt. For mouse dysbindin, only the largest isoform dysbindin-1A (NP_080048) is listed in NCBI Entrez Protein database and three in UniProt. Because up to 16 alternative spliced transcripts of DTNBP1 have been identified in human,31 it is possible there are more isoform proteins being translated that have not yet been detected.
To isolate the dysbindin-containing complexes from P2, we designed a tagging method according to the tandem affinity tag (TAP)-based strategy that has been shown to be very useful for purifying in vivo/endogenous protein complexes.64 Unlike the original method done in yeast Saccharomyces cerevisiae, tagging of the protein of interest directly on its chromosome locus in mice is more difficult and time-consuming. The BAC transgenic method has emerged as an alternative based on gene manipulation on BAC constructs in E. coli using recombineering.42 One advantage of using the BAC transgene compared with the commonly used cDNA overexpression method is that the BAC transgenes are much larger and generated from genomic DNAs, which include most, if not all, regulatory elements, such as native promoters. The result is expression of products similar to that from the endogenous gene. In addition, the intron-containing BACs can undergo alternative splicing and produce tagged isoforms, except in the case of an alternative first or last exon. For this reason, we generated both N- and C-terminal tagged dysbindin in BACs. However, we failed to obtain BAC transgenic mice that produced C-terminal tagged dysbindin. Although we successfully generated the BAC transgenic mice expressing the N-terminal tagged dysbindin, to our surprise, only one tagged dysbindin isoform (dysbindin-1A) could be detected. According to the characterized human dysbindin isoforms, the A and B isoforms contain the same N-terminal end, whereas A and C possess the same C-terminal end.24 UniProt database lists 3 mouse dysbindin isoforms, and based on the amino acid sequence alignment, the A and B isoforms contain the same N-terminal end, whereas all 3 isoforms possess the same C-terminal end. Therefore, we expected that both the N-terminal tagged dysbindin-1A and -1B would be expressed in the BAC transgenic mice. If the normal splicing is sensitive to the manipulation of the dysbindin genomic sequence or the splicing occurs at exons on both ends, then future analysis with the cDNA-based method may ensure isoform expression in mice.
Our mass spectrometric analysis led to identification of 47 putative dysbindin-associated proteins in the P2 fraction of mouse brain, including trafficking/transport, signaling, ubiquitin-proteasome system, transcription regulation, cell adhesion, and lipid metabolism related proteins. The identification of the whole BLOC-1 complex not only confirms that dysbindin is part of BLOC-1 in brain but also validates our proteomics analysis, as BLOC-1 subunits, pallidin and snapin, were previously shown to be associated with dysbindin in the brain.29,30 To our knowledge, this is the first systematic proteomics survey of the endogenous dysbindin-associated proteins in mouse brain. Previously, an affinity chromatography-based PPI study was performed for the dysbindin in rat brain using the purified GST-tagged dysbindin as a bait to isolate dysbindin-binding proteins from the cytosol and P2 fraction.65 Only dynamin-3 (DNM3) was identified in both the present and the previous analyses. The poor overlap between these two studies implies that protein complexes formed in vivo may not be able to be rearranged/reformed in vitro. In addition to BLOC-1, AP-3 complex subunit beta-1 (AP3B1), exocyst complex component 3 (EXOC3), and exocyst complex component 4 (EXOC4), identified in our study, were also reported previously from proteomics analyses using cell lines.38,39
More than two-thirds of the identified proteins in this study have not been reported previously. Although the verification and functional analysis of each protein on our list are beyond the scope of the current study, we were able to confirm several novel interacting proteins of dysbindin, including FAM91A1, WDR11, PSMD9, and PSMA4. The biological function of FAM91A1 is largely unknown. WDR11 was identified as a gene involved in human puberty. Mutations in WDR11 are associated with idiopathic hypogonadotropic hypogonadism and Kallmann syndrome.66 PSMD9 and PSMA4 are two subunits of 26S proteasome. Because dysbindin can be ubiquitinated,59 the association between dysbindin and proteasome subunits suggests that dysbindin could potentially be recruited to the proteasome for degradation through these interactions. Our proteasomal activity tests in the brains of sandy mice also suggest that dysbindin may play a role in the regulation of proteasomal activity. Therefore, the overall evidence suggests that dysbindin is functionally interrelated to the UPS, as it can serve as both substrate and regulator of the UPS. Because UPS is important for synaptic function, neuronal development, protein homeostasis, and other cellular processes in neurons, our data suggest a new functional linkage between dysbindin and SCZ. Future studies focusing on the novel candidates identified in this study should advance our understanding of dysbindin and its role in SCZ.
Acknowledgments
Our sincere thanks to Jim Makusky, Ronald Finnegan (scientific applications and virtual machine services), Dr. Brian Martin, Dr. Michael Strader, Dr. Adele Blackler, Dr. Jeff Kowalak, Dr. Yan Leng, Dr. Song Jiao, and Dr. Jie-Min Jia at NIMH for technical support, advice, and assistance. This research was supported by the Intramural Research Program of the National Institute of Mental Health, project nos. 1ZIAMH000274-35, 1ZIAMH00279-27, and 1ZIAMH002881.
Supporting Information Available
The second network (Cell-Mediated Immune Response, Cellular Development, Cellular Function and Maintenance) identified by Ingenuity Pathway Analysis of the 48 identified proteins; the 47 putative dysbindin-associated proteins grouped into seven categories with regard to their functions based on the information from UniProt, NCBI, and/or Gene Ontology databases. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Tandon R.; Keshavan M. S.; Nasrallah H. A. Schizophrenia, “just the facts” what we know in 2008. 2. Epidemiology and etiology. Schizophr. Res. 2008, 102, 1–18. [DOI] [PubMed] [Google Scholar]
- Sullivan P. F.; Kendler K. S.; Neale M. C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 2003, 60, 1187–92. [DOI] [PubMed] [Google Scholar]
- Sullivan P. F. Schizophrenia genetics: the search for a hard lead. Curr. Opin Psychiatry 2008, 21, 157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno A. G.; Gottesman I. I. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am. J. Med. Genet. 2000, 97, 12–7. [PubMed] [Google Scholar]
- O’Tuathaigh C. M.; Babovic D.; O’Meara G.; Clifford J. J.; Croke D. T.; Waddington J. L. Susceptibility genes for schizophrenia: characterisation of mutant mouse models at the level of phenotypic behaviour. Neurosci. Biobehav. Rev. 2007, 31, 60–78. [DOI] [PubMed] [Google Scholar]
- Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008, 455, 237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub R. E.; Jiang Y.; MacLean C. J.; Ma Y.; Webb B. T.; Myakishev M. V.; Harris-Kerr C.; Wormley B.; Sadek H.; Kadambi B.; Cesare A. J.; Gibberman A.; Wang X.; O’Neill F. A.; Walsh D.; Kendler K. S. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 2002, 71, 337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. M.; Moran J. L.; Fromer M.; Ruderfer D.; Solovieff N.; Roussos P.; O’Dushlaine C.; Chambert K.; Bergen S. E.; Kahler A.; Duncan L.; Stahl E.; Genovese G.; Fernandez E.; Collins M. O.; Komiyama N. H.; Choudhary J. S.; Magnusson P. K.; Banks E.; Shakir K.; Garimella K.; Fennell T.; DePristo M.; Grant S. G.; Haggarty S. J.; Gabriel S.; Scolnick E. M.; Lander E. S.; Hultman C. M.; Sullivan P. F.; McCarroll S. A.; Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014, 506, 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. M.; Wray N. R.; Stone J. L.; Visscher P. M.; O’Donovan M. C.; Sullivan P. F.; Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460, 748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.; Kumar A.; Agarwal S.; Phadke S. R.; Jaiswal Y. Genetic insight of schizophrenia: past and future perspectives. Gene 2014, 535, 97–100. [DOI] [PubMed] [Google Scholar]
- Balu D. T.; Coyle J. T. Neuroplasticity signaling pathways linked to the pathophysiology of schizophrenia. Neurosci. Biobehav. Rev. 2011, 35, 848–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiani C. A.; Dell’Angelica E. C. Dysbindin-containing complexes and their proposed functions in brain: from zero to (too) many in a decade. ASN Neuro 2011, 3, 109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.; Kuo P. H.; Riley B. P.; Kendler K. S.; Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am. J. Med. Genet., Part B 2008, 147B, 1173–81. [DOI] [PubMed] [Google Scholar]
- Allen N. C.; Bagade S.; McQueen M. B.; Ioannidis J. P.; Kavvoura F. K.; Khoury M. J.; Tanzi R. E.; Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008, 40, 827–34. [DOI] [PubMed] [Google Scholar]
- Sanders A. R.; Duan J.; Levinson D. F.; Shi J.; He D.; Hou C.; Burrell G. J.; Rice J. P.; Nertney D. A.; Olincy A.; Rozic P.; Vinogradov S.; Buccola N. G.; Mowry B. J.; Freedman R.; Amin F.; Black D. W.; Silverman J. M.; Byerley W. F.; Crowe R. R.; Cloninger C. R.; Martinez M.; Gejman P. V. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am. J. Psychiatry 2008, 165, 497–506. [DOI] [PubMed] [Google Scholar]
- Joo E. J.; Lee K. Y.; Jeong S. H.; Ahn Y. M.; Koo Y. J.; Kim Y. S. The dysbindin gene (DTNBP1) and schizophrenia: no support for an association in the Korean population. Neurosci. Lett. 2006, 407, 101–6. [DOI] [PubMed] [Google Scholar]
- Strohmaier J.; Frank J.; Wendland J. R.; Schumacher J.; Jamra R. A.; Treutlein J.; Nieratschker V.; Breuer R.; Mattheisen M.; Herms S.; Muhleisen T. W.; Maier W.; Nothen M. M.; Cichon S.; Rietschel M.; Schulze T. G. A reappraisal of the association between dysbindin (DTNBP1) and schizophrenia in a large combined case-control and family-based sample of German ancestry. Schizophr. Res. 2010, 118, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgenes K. V.; Djurovic S.; Agartz I.; Jonsson E. G.; Werge T.; Melle I.; Andreassen O. A. Dysbindin and d-amino-acid-oxidase gene polymorphisms associated with positive and negative symptoms in schizophrenia. Neuropsychobiology 2009, 60, 31–6. [DOI] [PubMed] [Google Scholar]
- DeRosse P.; Funke B.; Burdick K. E.; Lencz T.; Ekholm J. M.; Kane J. M.; Kucherlapati R.; Malhotra A. K. Dysbindin genotype and negative symptoms in schizophrenia. Am. J. Psychiatry 2006, 163, 532–4. [DOI] [PubMed] [Google Scholar]
- Pae C. U.; Drago A.; Kim J. J.; Patkar A. A.; Jun T. Y.; Lee C.; Mandelli L.; De Ronchi D.; Paik I. H.; Serretti A. DTNBP1 haplotype influences baseline assessment scores of schizophrenic in-patients. Neurosci. Lett. 2008, 440, 150–4. [DOI] [PubMed] [Google Scholar]
- Talbot K.; Eidem W. L.; Tinsley C. L.; Benson M. A.; Thompson E. W.; Smith R. J.; Hahn C. G.; Siegel S. J.; Trojanowski J. Q.; Gur R. E.; Blake D. J.; Arnold S. E. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Invest. 2004, 113, 1353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert C. S.; Straub R. E.; McClintock B. W.; Matsumoto M.; Hashimoto R.; Hyde T. M.; Herman M. M.; Weinberger D. R.; Kleinman J. E. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch. Gen. Psychiatry 2004, 61, 544–55. [DOI] [PubMed] [Google Scholar]
- Talbot K.; Louneva N.; Cohen J. W.; Kazi H.; Blake D. J.; Arnold S. E. Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS One 2011, 6, e16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.; LeGros R. P.; Louneva N.; Yeh L.; Cohen J. W.; Hahn C. G.; Blake D. J.; Arnold S. E.; Talbot K. Dysbindin-1 in dorsolateral prefrontal cortex of schizophrenia cases is reduced in an isoform-specific manner unrelated to dysbindin-1 mRNA expression. Hum. Mol. Genet. 2009, 18, 3851–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. J.; Preece A.; Williams N. M.; Moskvina V.; Buckland P. R.; Owen M. J.; O’Donovan M. C. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum. Mol. Genet. 2005, 14, 1947–54. [DOI] [PubMed] [Google Scholar]
- Feng Y. Q.; Zhou Z. Y.; He X.; Wang H.; Guo X. L.; Hao C. J.; Guo Y.; Zhen X. C.; Li W. Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr. Res. 2008, 106, 218–28. [DOI] [PubMed] [Google Scholar]
- Talbot K. The sandy (sdy) mouse: a dysbindin-1 mutant relevant to schizophrenia research. Prog. Brain Res. 2009, 179, 87–94. [DOI] [PubMed] [Google Scholar]
- Bhardwaj S. K.; Baharnoori M.; Sharif-Askari B.; Kamath A.; Williams S.; Srivastava L. K. Behavioral characterization of dysbindin-1 deficient sandy mice. Behav. Brain Res. 2009, 197, 435–41. [DOI] [PubMed] [Google Scholar]
- Benson M. A.; Newey S. E.; Martin-Rendon E.; Hawkes R.; Blake D. J. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 2001, 276, 24232–41. [DOI] [PubMed] [Google Scholar]
- Talbot K.; Cho D. S.; Ong W. Y.; Benson M. A.; Han L. Y.; Kazi H. A.; Kamins J.; Hahn C. G.; Blake D. J.; Arnold S. E. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum. Mol. Genet. 2006, 15, 3041–54. [DOI] [PubMed] [Google Scholar]
- Guo A. Y.; Sun J.; Riley B. P.; Thiselton D. L.; Kendler K. S.; Zhao Z. The dystrobrevin-binding protein 1 gene: features and networks. Mol. Psychiatry 2009, 14, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiani C. A.; Starcevic M.; Rodriguez-Fernandez I. A.; Nazarian R.; Cheli V. T.; Chan L. N.; Malvar J. S.; de Vellis J.; Sabatti C.; Dell’Angelica E. C. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol. Psychiatry 2010, 15, 115.204–15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankle W. G.; Lerma J.; Laruelle M. The synaptic hypothesis of schizophrenia. Neuron 2003, 39, 205–16. [DOI] [PubMed] [Google Scholar]
- Li W.; Zhang Q.; Oiso N.; Novak E. K.; Gautam R.; O’Brien E. P.; Tinsley C. L.; Blake D. J.; Spritz R. A.; Copeland N. G.; Jenkins N. A.; Amato D.; Roe B. A.; Starcevic M.; Dell’Angelica E. C.; Elliott R. W.; Mishra V.; Kingsmore S. F.; Paylor R. E.; Swank R. T. Hermansky–Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 2003, 35, 84–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen G.; Prata D.; Osborne S.; Munro J.; Sinclair M.; Li T.; Staddon S.; Dempster D.; Sainz R.; Arroyo B.; Kerwin R. W.; St. Clair D.; Collier D. Association of the dysbindin gene with bipolar affective disorder. Am. J. Psychiatry 2006, 163, 1636–8. [DOI] [PubMed] [Google Scholar]
- Sardiu M. E.; Washburn M. P. Building protein–protein interaction networks with proteomics and informatics tools. J. Biol. Chem. 2011, 286, 23645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo L. M.; Collura V.; Rain J. C.; Mizuguchi K.; Hermjakob H.; Kerrien S.; Bonnert T. P.; Whiting P. J.; Brandon N. J. Disrupted in Schizophrenia 1 interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry 2007, 12, 74–86. [DOI] [PubMed] [Google Scholar]
- Mead C. L.; Kuzyk M. A.; Moradian A.; Wilson G. M.; Holt R. A.; Morin G. B. Cytosolic protein interactions of the schizophrenia susceptibility gene dysbindin. J. Neurochem 2010, 113, 1491–503. [DOI] [PubMed] [Google Scholar]
- Gokhale A.; Larimore J.; Werner E.; So L.; Moreno-De-Luca A.; Lese-Martin C.; Lupashin V. V.; Smith Y.; Faundez V. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J. Neurosci. 2012, 32, 3697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitre S.; Alamgir M.; Green J. R.; Dumontier M.; Dehne F.; Golshani A. Computational methods for predicting protein-protein interactions. Adv. Biochem. Eng. Biotechnol. 2008, 110, 247–67. [DOI] [PubMed] [Google Scholar]
- Whittaker V. P. Thirty years of synaptosome research. J. Neurocytol. 1993, 22, 735–42. [DOI] [PubMed] [Google Scholar]
- Poser I.; Sarov M.; Hutchins J. R.; Heriche J. K.; Toyoda Y.; Pozniakovsky A.; Weigl D.; Nitzsche A.; Hegemann B.; Bird A. W.; Pelletier L.; Kittler R.; Hua S.; Naumann R.; Augsburg M.; Sykora M. M.; Hofemeister H.; Zhang Y.; Nasmyth K.; White K. P.; Dietzel S.; Mechtler K.; Durbin R.; Stewart A. F.; Peters J. M.; Buchholz F.; Hyman A. A. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 2008, 5, 409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S.; Costantino N.; Court D. L.; Jenkins N. A.; Copeland N. G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005, 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M. H.; Lin C.; Meng S.; Wang X. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol. Cell. Proteomics 2010, 9, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A.; Nesvizhskii A. I.; Kolker E.; Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–92. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Keller A.; Kolker E.; Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–58. [DOI] [PubMed] [Google Scholar]
- Han M. H.; Jiao S.; Jia J. M.; Chen Y.; Chen C. Y.; Gucek M.; Markey S. P.; Li Z. The novel caspase-3 substrate Gap43 is involved in AMPA receptor endocytosis and long-term depression. Mol. Cell. Proteomics 2013, 12, 3719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucksberg K. H.; Tangavelou K.; Schroder R.; Clemen C. S. Proteasomal activity in skeletal muscle: a matter of assay design, muscle type, and age. Anal. Biochem. 2010, 399, 225–9. [DOI] [PubMed] [Google Scholar]
- Salazar G.; Zlatic S.; Craige B.; Peden A. A.; Pohl J.; Faundez V. Hermansky–Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J. Biol. Chem. 2009, 284, 1790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Rayner S.; Katoku-Kikyo N.; Romanova L.; Kikyo N. Successful co-immunoprecipitation of Oct4 and Nanog using cross-linking. Biochem. Biophys. Res. Commun. 2007, 361, 611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon-Perez J. M.; Starcevic M.; Gautam R.; Dell’Angelica E. C. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 2002, 277, 28191–9. [DOI] [PubMed] [Google Scholar]
- Hollenback S. M.; Lyman S.; Cheng J.. Recombineering-based procedure for creating BAC transgene constructs for animals and cell lines. Current Protocols in Molecular Biology; John Wiley & Sons: New York, 2011; Chapter 23, Unit 23 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keuren M. L.; Gavrilina G. B.; Filipiak W. E.; Zeidler M. G.; Saunders T. L. Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 2009, 18, 769–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler K. J.; Chandler R. L.; Broeckelmann E. M.; Hou Y.; Southard-Smith E. M.; Mortlock D. P. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm. Genome 2007, 18, 693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R.; Starcevic M.; Spencer M. J.; Dell’Angelica E. C. Reinvestigation of the dysbindin subunit of BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem. J. 2006, 395, 587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. L.; Dong C.; Andreev V.; Arcos-Burgos M.; Licinio J. Prediction of susceptibility to major depression by a model of interactions of multiple functional genetic variants and environmental factors. Mol. Psychiatry 2012, 17, 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragnoli C. Proteasome modulator 9 and depression in type 2 diabetes. Curr. Med. Chem. 2012, 19, 5178–5180. [DOI] [PubMed] [Google Scholar]
- Lee Y. H.; Kim J. H.; Song G. G. Pathway analysis of a genome-wide association study in schizophrenia. Gene 2013, 525, 107–15. [DOI] [PubMed] [Google Scholar]
- Locke M.; Tinsley C. L.; Benson M. A.; Blake D. J. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009, 18, 2344–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome T. J.; Helmstetter F. J. The ubiquitin-proteasome system as a critical regulator of synaptic plasticity and long-term memory formation. Neurobiol. Learn. Mem. 2013, 105, 107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A. W.; Man H. Y. Ubiquitination of neurotransmitter receptors and postsynaptic scaffolding proteins. Neural Plast. 2013, 2013, 432057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb A. M.; Ehlers M. D. Ubiquitination in postsynaptic function and plasticity. Annu. Rev. Cell Dev. Biol. 2010, 26, 179–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H. C.; Besche H.; Goldberg A. L.; Schuman E. M. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol. Neurosci. 2010, 3, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G.; Shevchenko A.; Rutz B.; Wilm M.; Mann M.; Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999, 17, 1030–2. [DOI] [PubMed] [Google Scholar]
- Hikita T.; Taya S.; Fujino Y.; Taneichi-Kuroda S.; Ohta K.; Tsuboi D.; Shinoda T.; Kuroda K.; Funahashi Y.; Uraguchi-Asaki J.; Hashimoto R.; Kaibuchi K. Proteomic analysis reveals novel binding partners of dysbindin, a schizophrenia-related protein. J. Neurochem. 2009, 110, 1567–74. [DOI] [PubMed] [Google Scholar]
- Kim H. G.; Ahn J. W.; Kurth I.; Ullmann R.; Kim H. T.; Kulharya A.; Ha K. S.; Itokawa Y.; Meliciani I.; Wenzel W.; Lee D.; Rosenberger G.; Ozata M.; Bick D. P.; Sherins R. J.; Nagase T.; Tekin M.; Kim S. H.; Kim C. H.; Ropers H. H.; Gusella J. F.; Kalscheuer V.; Choi C. Y.; Layman L. C. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 2010, 87, 465–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.