Figure 2.
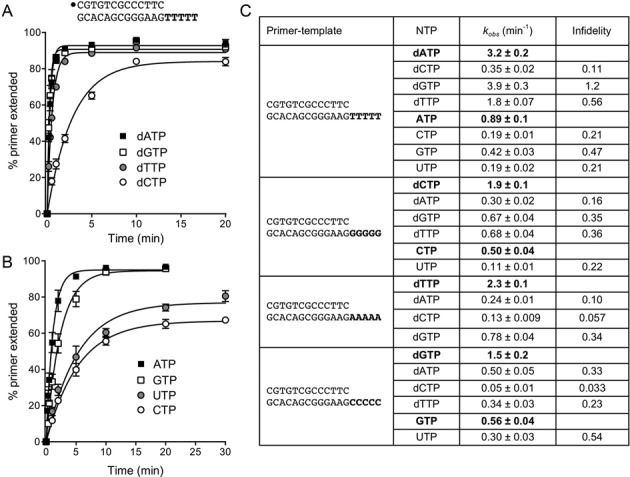
Kinetic analysis of DinB2 infidelity with manganese as the metal cofactor. (A and B) Polymerase reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 50 nM 5′ 32P-labeled 13-mer/18-mer primer-template with an oligo-dT5 tail, 1 mM MnCl2, and either 100 μM deoxynucleoside triphosphate (panel A) or 100 μM ribonucleoside triphosphate (panel B) as specified were incubated at 37°C. Aliquots (10 μl) were withdrawn at the times specified and quenched immediately with EDTA/formamide. The reaction products were analyzed by urea-PAGE and the percent of primer strand extended by one or more nucleotides was quantified by scanning the gel with a Fujix BAS2500 imager. The % primer extension is plotted as a function of reaction time. Each datum in the graphs is the average of three separate experiments ±SEM. The rate constants (kobs ± SE) for the first step of dNMP and rNMP addition to the primer-template were obtained by non-linear regression curve fitting of the data to a one-phase association function in Prism and are shown in panel C. A series of 5′ 32P-labeled 13-mer/18-mer primer-templates with identical duplex segments and either oligo-dG5, oligo-dA5 or oligo-dC5 template tails was used to assay the kinetics of nucleotide addition to the primer strand. The rate constants are compiled in panel C. Infidelity was calculated as the ratio of the rate of mispaired deoxynucleotide or ribonucleotide addition to the rate of addition of the correctly paired deoxynucleotide or ribonucleotide.
