Figure 3.
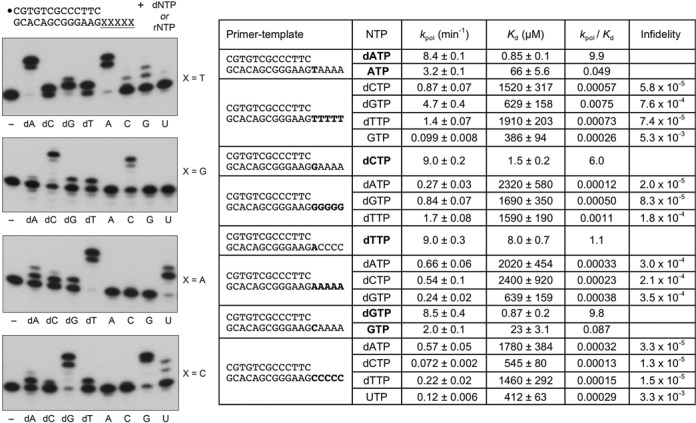
dNMP and rNMP misincorporation during magnesium-dependent synthesis. Left panel. Polymerase reaction mixtures (10 μl) containing 10 mM Tris-HCl, pH 7.5, 50 nM 5′ 32P-labeled 13-mer/18-mer primer-template as specified, 1 μM DinB2, 5 mM MgCl2 and either no nucleotide (–) or 500 μM of the indicated rNTP or dNTP were incubated at 37°C for 10 min. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. Right panel. Polymerase reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 nM 5′ 32P-labeled 13-mer/18-mer primer-template, 1 μM DinB2, and varying concentrations of dNTP or rNTP substrates were incubated at 37°C. Aliquots (10 μl) were withdrawn at sequential timepoints and quenched with EDTA/formamide. The products were analyzed by urea-PAGE and quantified by scanning the gels. The % of primer extended by one or more nucleotides was plotted as a function of reaction time for each concentration of dNTP or rNTP substrate. The data were fit by non-linear regression in Prism to a one-phase exponential. The kobs values calculated in Prism were plotted as a function of ATP concentration, with each datum being the average of two separate kinetic experiments. The data were fit by non-linear regression to a single binding function, from which the Kd for nucleotide and turnover number (kpol) at saturating nucleotide were derived. The kpol and Kd values and kpol/Kd ratio (catalytic efficiency) for each dNTP and rNTP substrate are indicated. Infidelity was calculated as the ratio of the catalytic efficiency for the mispaired versus correctly paired deoxynucleotide and ribonucleotide substrates.
