Figure 5.
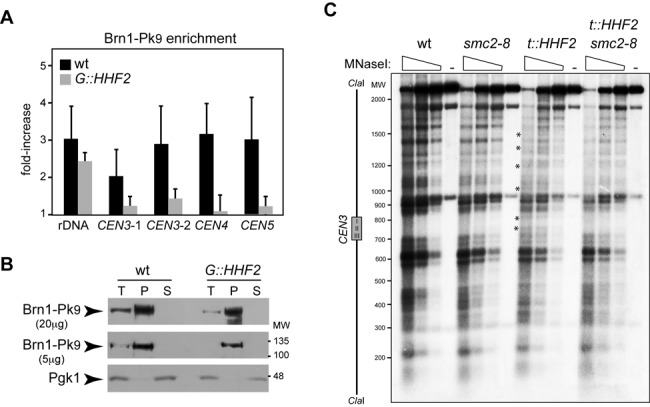
Interplay between histone supply and condensin. (A) Condensin enrichment at different positions of the pericentric chromatin and the ribosomal DNA by ChIP analysis of Brn1-Pk9 in wild-type and G::HHF2 cells grown as indicated in Figure 4D. The analyzed regions lie at the 5′ untranslated region of RDN37 (rDNA) and at 3.35 (CEN3-1), 1.74 (CEN3-2), 0.5 (CEN4) and 2.6 kb (CEN5) from the corresponding centromere. The average and SEM of three independent experiments are shown. The differences are not statistically significant. (B) Western analysis of Brn1 after chromatin fractionation of wild-type and G::HHF2 cells grown as indicated in Figure 4D. Twenty (upper panel) or five micrograms (middle panel) of total (T), pellet (P) and soluble (S) proteins were analyzed. Pgk1 was used as loading control (lower panel). (C) Defective centromeric chromatin structure by histone depletion depends on condensin. MNaseI digestion and indirect-end labeling of CEN3 in wild-type, smc2-8, t::HHF2 and t::HHF2 smc2-8 cells synchronized in G1 and released into fresh medium at restrictive temperature (37°C) until G2/M (90 min for wild-type and smc2-8 and 120 min for t::HHF2 and t::HHF2 smc2-8) under conditions of histone depletion. A scheme with the position of the CDE I-II-III sequence of CEN3 is shown on the left. Asterisks show sites of hypersensitivity to MNaseI. The experiment was repeated twice with similar results.
