Figure 1.
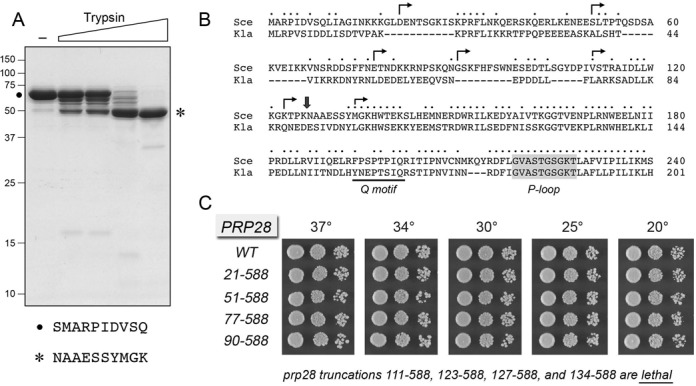
Limited proteolysis of Prp28 and effects of N-terminal truncations on Prp28 activity in vivo. (A) Limited proteolysis. Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 10 mM MgCl2, 5 μg of full-length Prp28 and increasing amounts of trypsin (0, 1, 2, 10, 50 ng, from left to right) were incubated for 15 min at room temperature. The mixtures were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. The N-terminal sequences of the intact Prp28 polypeptide (•) and the 50-kDa tryptic fragment (asterisk) are shown below the gel. (B) The amino acid sequence of the N-terminal segment of S. cerevisiae (Sce) Prp28 upstream of Q motif (bracketed) and the P-loop (shaded grey) is aligned to that of the homologous protein from Kluyveromyces lactis (Kla). Positions of amino acid site chain identity/similarity are denoted by • above the sequence. Arrowheads indicate the boundaries of the N-terminal deletions of Prp28. (C) The wild-type and deleted PRP28 alleles were tested for activity by plasmid shuffle as described under Materials and Methods. The growth phenotypes of viable FOA-resistant prp28Δ p[CEN HIS3 PRP28] strains bearing the indicated PRP28 alleles were assessed as follows. Liquid cultures were grown to mid-log phase and adjusted to the same A600. Aliquots (3 μl) of serial 10-fold dilutions of cells were spotted to YPD agar. The plates were incubated at the indicated temperatures and photographed after 2 days (30, 34 and 37°C), 3 days (25°C) or 4 days (20°C). The truncated mutants listed at bottom failed to complement prp28Δ in a plasmid shuffle assay and were deemed lethal.
