Figure 2.
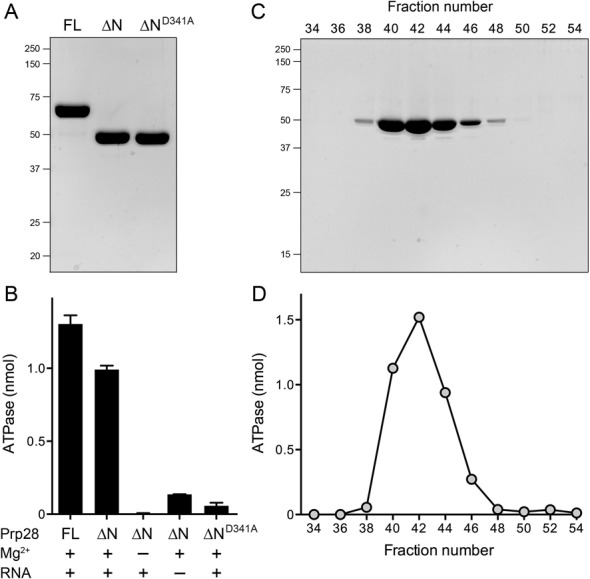
RNA-dependent ATPase activity. (A) Recombinant proteins. Aliquots (5 μg) of the Superdex-200 preparations of full-length Prp28 (FL), Prp28-(127–588) (NΔ) and Prp28-(127–588)-D341A (NΔD341A) were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) ATPase reaction mixtures (10 μl) containing 20 mM Tris-HCl, pH 7.1, 1 mM (10 nmol) [α32P]ATP, 10 mM MgCl2 (where indicated by +), poly(U) RNA (200 μM total uridine nucleotide, where indicated by +) and 10 μM (100 pmol) of the indicated Prp28 protein were incubated at 30°C for 60 min. The reactions were quenched by adding 2 μl of 5 M formic acid. An aliquot of the mixture was spotted on a polyethyleneimine-cellulose thin layer chromatography (TLC) plate. Ascending TLC was performed with 0.45 M ammonium sulfate as the mobile phase. [α32P]ADP formation was quantified by scanning the TLC plate with a Fujix BAS2500 imager. Each datum in the graph is an average of three separate experiments ± SEM. (C) Gel filtration of Prp28-(127–588). Aliquots (1 μl) of the indicated even-numbered Superdex-200 fractions across the elution peak were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (D) Aliquots (1 μl) of the indicated Superdex-200 fractions from panel C were assayed for ATP hydrolysis in reaction mixtures constituted as described in panel B. The ATPase activity profile coincides with the elution profile of the Prp28-(127–588) polypeptide.
