Figure 5.
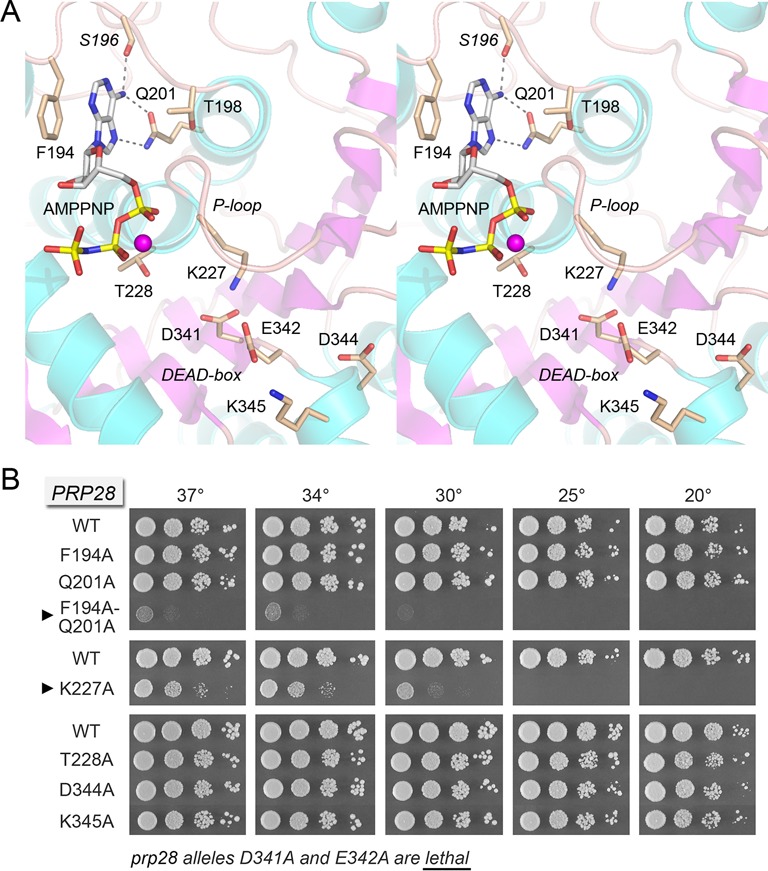
Prp28 binding to AMPPNP and mutational analysis of the binding site. (A) Stereo view of the ATP site in the N domain of the yeast Prp28-ΔN B protomer. Amino acids and AMPPNP are shown as stick models with beige and gray carbons, respectively. Amino acid side chains are labeled by single letter code and numbered; the S196 main chain is in italics. Mg2+ is depicted as a magenta sphere. The P-loop and DEAD-box motifs are indicated. Hydrogen bonds to the adenine N6 and N7 atoms are denoted by dashed lines. (B) The indicated PRP28 mutant alleles were tested for prp28Δ complementation by plasmid shuffle. D341A and E342A were lethal. The viable FOA-resistant strains were spot-tested for growth on YPD agar at the temperatures specified, in parallel with the isogenic wild-type PRP28 strain. Alleles with strong cold-sensitive growth defects are indicated by ▸ at left.
