Figure 6.
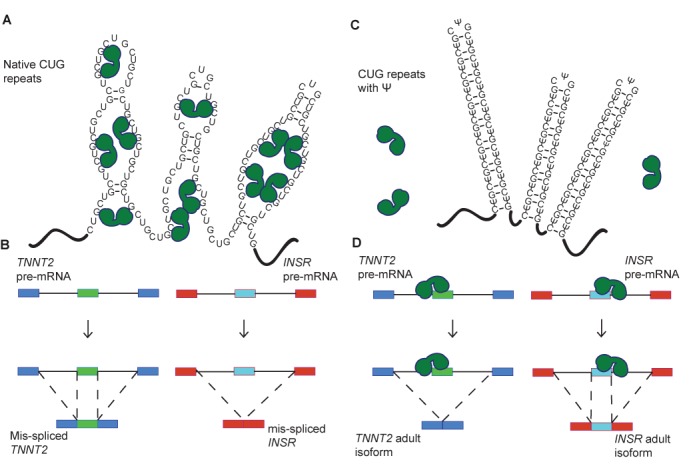
A model for CUG repeat detoxification via pseudouridylation. (A) Native CUG repeats form unstable A-form helices that are proposed to adopt multiple conformations, including partially unstructured RNA. Partially unfolded CUG repeats present the Watson–Crick faces of numerous MBNL1 binding sites (YGCY). In this case, MBNL1 binds to the CUG repeats and is sequestered to their location in the nucleus. MBNL1 is represented by green shapes. (B) When MBNL1 is sequestered to CUG repeats, it is unavailable to bind to the pre-mRNAs that it regulates. Here CUG repeats increased exon inclusion in the TNNT2 transcript, which is characteristic of the fetal isoform. CUG repeats result in decreased exon inclusion in the INSR mRNA in the presence of CUG repeats, also characteristic of the fetal isoform. (C) CUG repeats with Ψ substitutions (CΨG repeats) form stable stem-loop structures. In these structures the Watson–Crick face of MBNL1′s binding site is unavailable for interaction and MBNL1 is not sequestered. (D) MBNL1 is free to bind to its target transcripts when CΨG repeats are present, thus exon inclusion levels for the regulated exons in TNNT2 and INSR are wild-type.
