Figure 1.
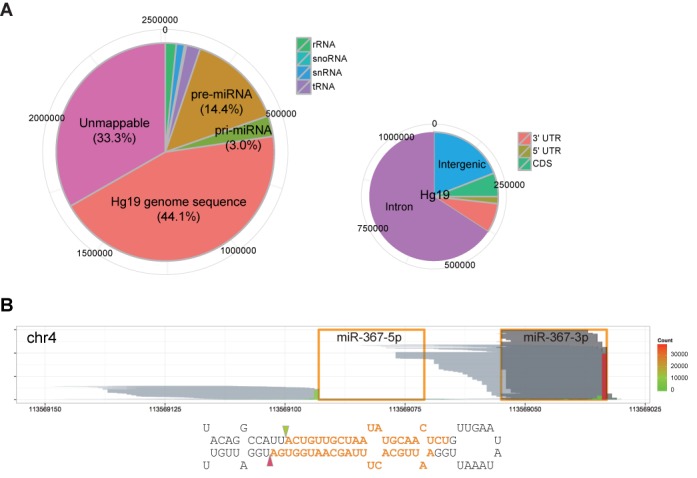
miRNA precursors are a major RNA class interacting with DGCR8. (A) Distribution of DGCR8-CLIP-seq reads on the genome. The larger pie chart shows percentages of collapsed reads mapping to non-coding RNAs and the UCSC human genome (hg19). The smaller pie chart shows distribution of reads uniquely mapping to the hg19 human genome in the intergenic, intronic, coding regions and UTRs. (B) Reads from the DGCR8-CLIP and AGO2-IP libraries aligned to the mir-367 locus. Orange line boxes and orange bold letters denote the annotated mature miR-367–5p and -3p in miRBase. Each pale gray horizontal line represents each collapsed read from the DGCR8-CLIP libraries. Positions of the vertical bars represent the positions of the 3′ terminal nucleotides of DGCR8-CLIP-seq reads, and heights and filled colors of the vertical bars represent counts of the 3′ terminal nucleotides of reads. Note that the pale gray lines map to the miR-367 precursors, but not to the mature miR-367, and the predominant 3′ termini of DGCR8-CLIP-seq reads, denoted by triangles, match the annotated cleavage sites in miRBase. Each dark gray horizontal line represents each read from the AGO2-IP libraries. For better visualization, 6000 reads are sampled from 338 890 AGO2-IP reads after mapping to the mir-367 locus. Note that the uneven 5′ ends of reads from the AGO2-IP libraries are caused by non-templated nucleotide addition to the 3′ end of cDNA.
