Figure 2.
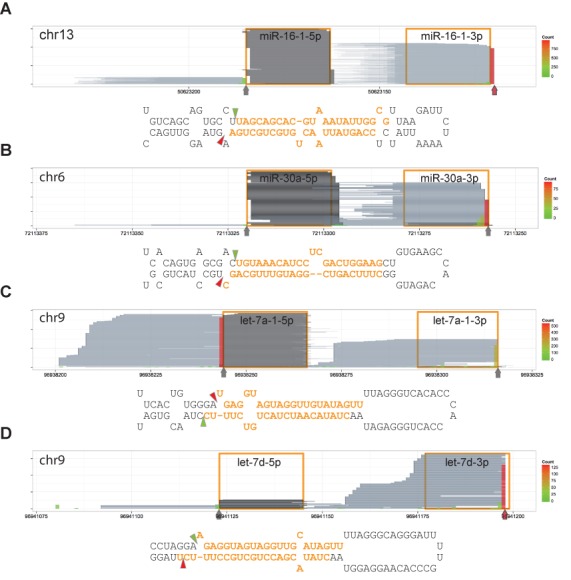
Major 3′ ends of reads from the DGCR8-CLIP libraries are identical to the in vitro cleavage sites. Reads are represented and denoted as the way in Figure 1B. Vertical arrows and triangles represent the predominant 3′ ends of DGCR8-CLIP reads. Predominant 3′ ends of reads that are different from annotated cleavage sites in miRBase are marked by red vertical arrows. (A) mir-16–1 locus. (B) mir-30a locus. (C) let-7a-1 locus. (D) let-7d locus. Note that the predominant 3′ ends of DGCR8-CLIP reads are identical to previously reported in vitro Microprocessor-mediated cleavage sites (25–27) rather than the annotations in miRBase.
