Figure 3.
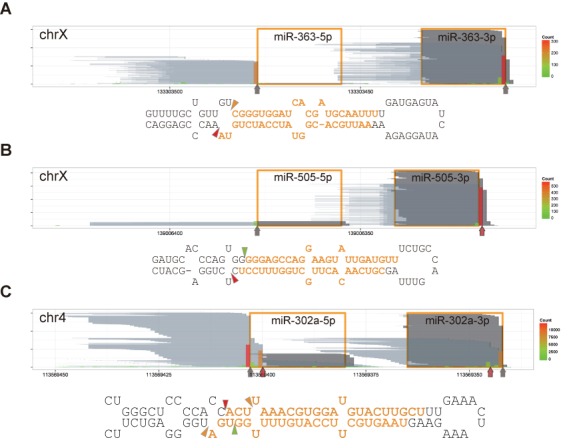
Non-templated nucleotide addition and alternative cleavage. Reads are represented as in Figure 1B. Vertical arrows and triangles indicate the major 3′ ends of DGCR8-CLIP-seq reads. Red vertical arrows mark the major 3′ ends of reads different from the Microprocessor-mediated cleavage site annotations or missing in miRBase. (A) Non-templated nucleotide addition to miR-363. Note that ∼50% of miR-363–3p from the AGO2-IP libraries have additional nucleotide(s) at the 3′ ends. (B) Non-templated nucleotide addition to miR-505. (C) Alternative cleavage sites of miR-302a. Individual short (up to 46 nt) and long (from 51 to 146 nt) reads of miR-302a precursors from the DGCR8-CLIP libraries are represented by pale gray and blue lines in Supplementary Figure S7, respectively. Note that 61- and 55-nt long reads correspond to predominant pre-miRNA isoforms, but miR-302a-5p is derived mainly from the 55-nt isoform, and miR-302a-3p is derived from the 61-nt isoform. Note the discrepancies between annotated miR-302a-5p and AGO2-associated miR-302a-5p.
