Figure 4.
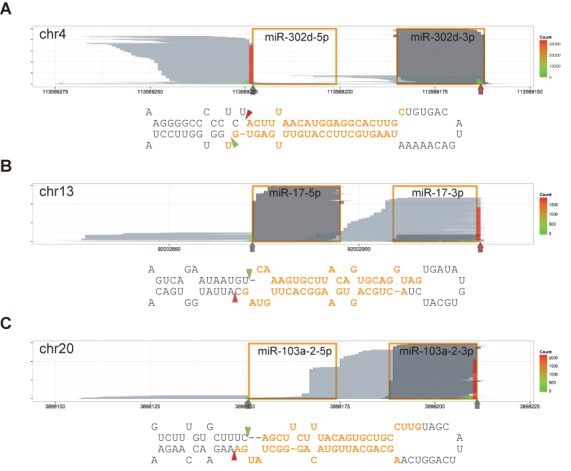
A 2-nt overhang of cleaved base is the hallmark of Microprocessor-mediated cleavage. Reads are represented as in Figure 1B and marked as in Figure 3. Predicted secondary structures are presented as in Figure 2. (A) DGCR8-CLIP-seq reads mapping to the mir-302d locus. Pri-miR-302d bears a 1-nt bulge in the 5′ arm. (B) DGCR8-CLIP-seq reads mapping to the mir-17 locus. Pri-miR-17 bears a 1-nt bulge in the 3′ arm. (C) DGCR8-CLIP-seq reads mapping to the mir-103a-2 locus. Pri-miR-103a-2 bears a 2-nt bulge in the 3′ arm. See also Supplementary Figure S10 for additional information on other bulged miRNAs. Note that the 5′ cleavage sites are located in the bulged structures, and that all the 3′ cleavage sites are in the non-bulged regions to release bases bearing a 2-nt 3′ overhang.
