Figure 5.
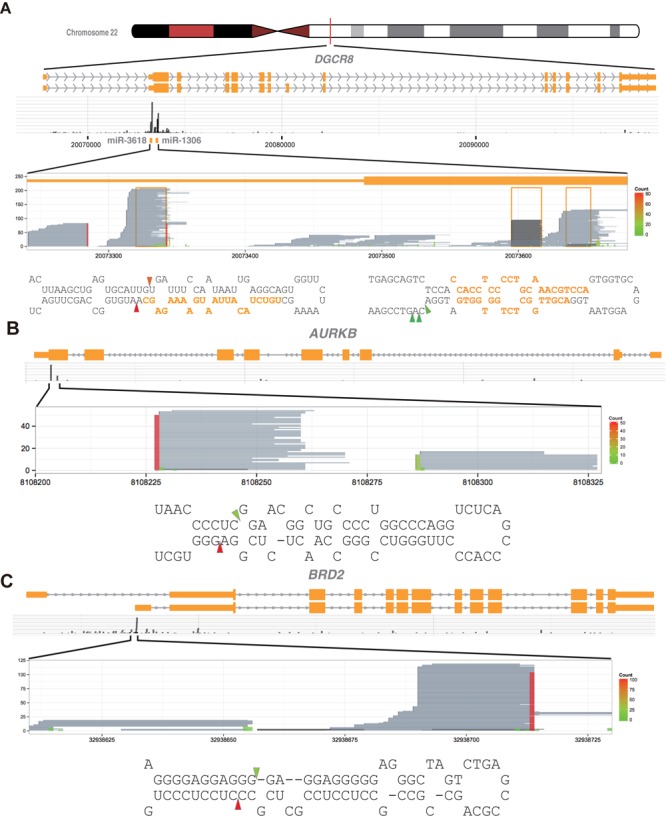
The Microprocessor cleaves miRNA-like structures embedded in mRNAs. (A–C) Shown are DGCR8-CLIP-enriched RNA fragments mapping to (A) the DGCR8, (B) the AURKB and (C) the BRD2 mRNAs. Reads aligned to the entire loci proclaim specificity of the DGCR8-CLIP experiments. Reads from the DGCR8-CLIP libraries and the Microprocessor-mediated cleavage sites in mRNAs are represented as in Figure 1B. Note that the stem-loop structures and cleavage sites have characteristics of typical pri-miRNAs, whereas the pre-miRNA-like hairpins are uncoupled from miRNA maturation.
