Figure 6.
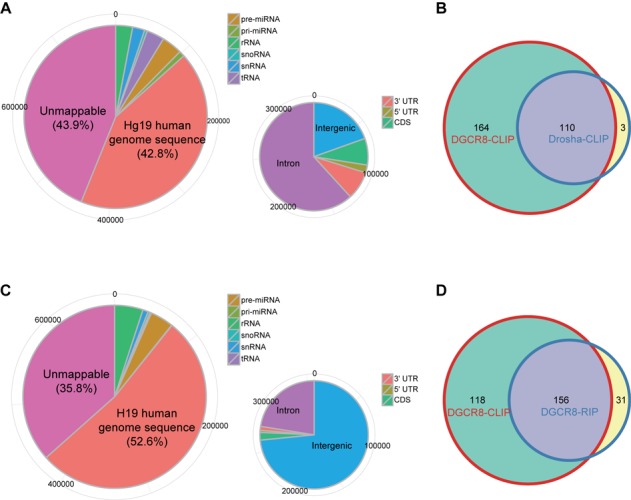
Features of CLIP-seq libraries for Drosha and a RIP-seq library for DGCR8. (A and C) Shown are distributions of locations of (A) Drosha-CLIP-seq and (B) DGCR8-RIP-seq reads on the human genome. Pie charts are presented as in Figure 1A. (B and D) Weighted Venn diagram of annotated miRNA precursors recovered by DGCR8-CLIP, (C) Drosha-CLIP and (D) DGCR8-RIP. Note that although we omit less frequently recovered miRNA precursors from DGCR8-CLIP (maximum coverage <8), miRNA precursors from DGCR8-CLIP overlap 110 (out of 113) and 156 (out of 187) miRNA precursors recovered from Drosha-CLIP and DGCR8-RIP, respectively.
