Figure 7.
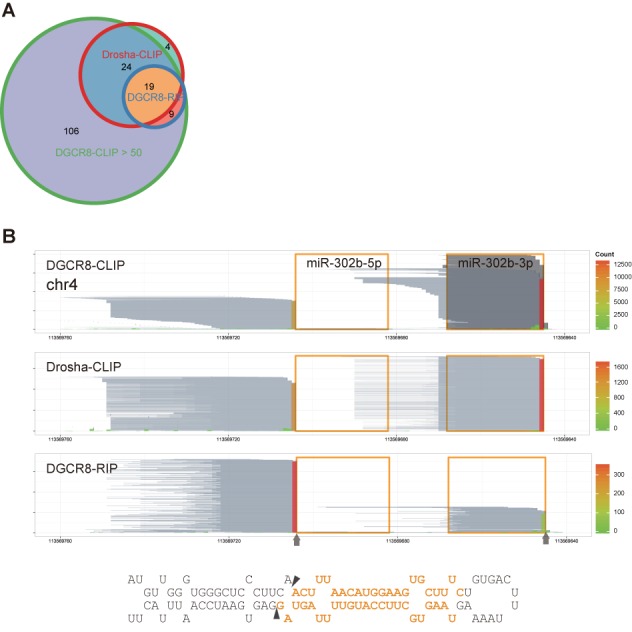
Cleavage sites determined by DGCR8-CLIP-seq reads are supported by Drosha-CLIP-seq and DGCR8-RIP-seq reads. (A) Weighted Venn diagram of cleavage sites on annotated miRNA precursors determined by the reads from DGCR8-CLIP (maximum coverage >50), Drosha-CLIP (>7) and DGCR8-RIP (>7). Cleavage sites determined by DGCR8-CLIP-seq are identical to the cleavages sites on 43 (out of 47) and 28 (out of 28) miRNA precursors determined by Drosha-CLIP-seq and DGCR8-RIP-seq, respectively. The four miRNAs uniquely recovered from Drosha-CLIP have low maximum coverage. (B) Reads from the DGCR8-CLIP, Drosha-CLIP and DGCR8-RIP libraries aligned to the mir-302b locus. Reads are represented as in Figure 1B and marked as in Figure 3. Note that the 5′ and 3′ cleavage sites determined from different experimental approaches are identical.
