Figure 1.
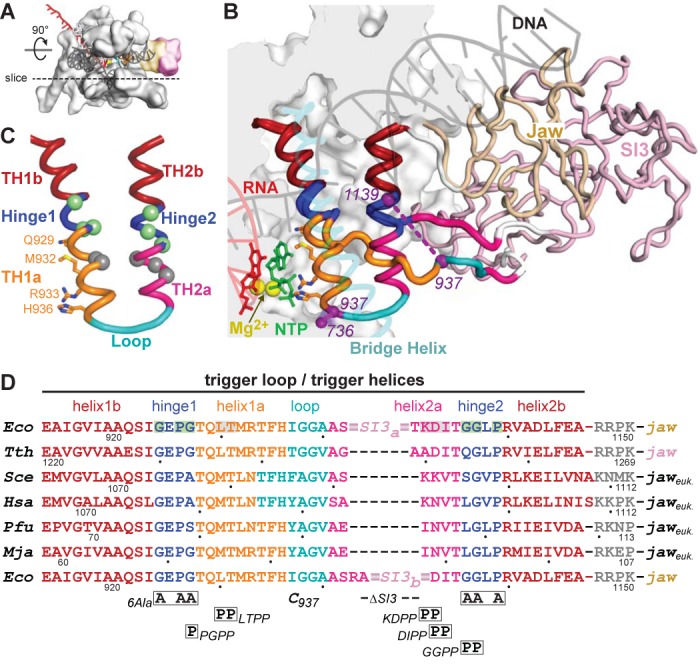
Structure and sequences of the TL/TH domain. (A) Model of the EcoRNAP EC in light gray space-fill showing the DNA (semi-transparent dark gray), RNA (red), active-site Mg2+ (yellow sphere) and the jaw (wheat) and SI3 (pink) domains. The dotted line depicts the approximate location of a slice through the EC that is rotated up ∼90° to give the main view in panel B. (B) Magnified view of TH and TL conformations in a model of the EcoRNAP EC based on the crystal structure of a TthRNAP EC bound to AMPCPP (PDB 2o5j (2) and a hydrid model of EcoRNAP (33)). The inset shows a space-filled rendering of EcoRNAP with some loops (BH, TL, lid, rudder, Zn-binding) shown as backbone worms for clarity. Nucleic acids, active-site Mg2+, jaw and SI3 domains are colored as in (A). The jaw (wheat) and SI3 (pink) domains are shown as worms, with only the connection of SI3 to the unfolded TL depicted for clarity (white segments are the connectors between the TL and SI3). The TH/TL is colored as follows: bases helices TH1b and TH2b (dark red), Hinges 1 and 2 (blue), TH1a (orange) and TH2a (magenta) and loop (cyan). The 188-aa SI3 is inserted between the loop and TH2a. Also shown are the incoming NTP (green), the BH (transparent cyan) and the Cys-pair reporters that detect the folded and unfolded conformation of the TH/TL (purple, F937–736 and U937–1139, respectively). Base helices TH1b and TH2b (dark red) do not change structure in the TL to TH transition. (C) Substructure of the TH and substitutions in EcoRNAP tested in this study. TH/TL is depicted in the same colors as in panel B also showing the positions of amino acids that make active site contacts to the NTP substrate to promote catalysis. The hinges (1 and 2; blue) contain Gly and Pro residues that destabilize the TH. Substitution locations depicted as spheres colored green (to stabilize) or gray (to destabilize) the TH, as highlighted in (D). (D) The sequences of the TL/TH in RNAPs from the three domains of life are shown colored as in panels B and C. Eco, Escherichia coli RNAP. Tth, Thermus thermophilus RNAP. Sce, Sacchaaromyces cerevisiae RNAPII. Hsa, Homo sapiens RNAPII. Pfu, Pyrococcus furiosis RNAP. Mja, Methanocaldococcus jannaschii RNAP. The locations of substitutions tested in this study are shown below the sequence alignment, with the two different SI3 deletions (SI3a and SI3b) shown in the top and bottom rows, respectively. The eukaryotic and archaeal jaw domains are not SBHM folds like the bacterial jaw domains.
