Figure 1.
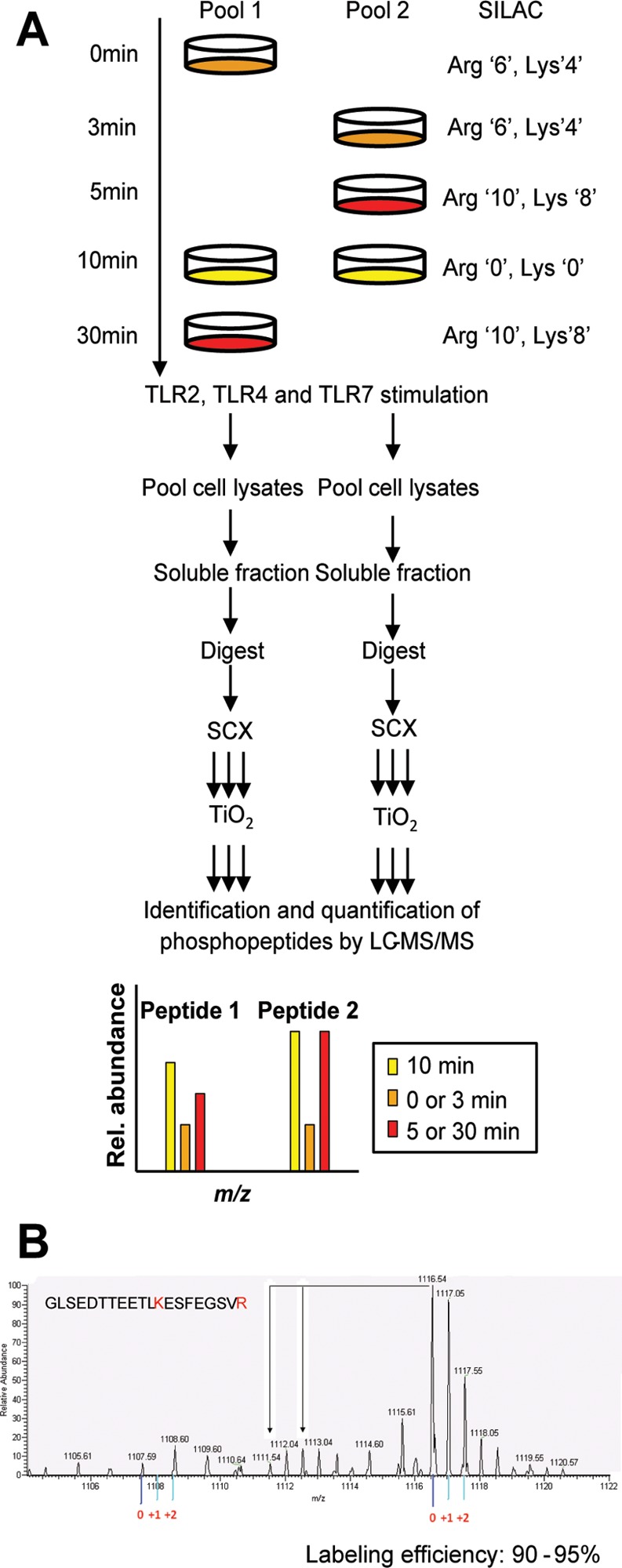
Experimental system and design. (A) Strategy for global and quantitative analyses of LPS-, P3C-, and R848-induced phosphorylation. C57 derived macrophages were SILAC labeled with normal or stable isotope-substituted arginine and lysine amino acids resulting in three states distinguishable by their mass. Each cell population was left untreated or stimulated for 3, 5, 10, or 30 min. The 10 min stimulation time point was included in both pools to serve as a common reference point. Cell lysates to be directly compared were pooled, enzymatically digested, and fractionated by SCX. The phosphopeptides were enriched by TiO2 and analyzed by online LC–MS(/MS). The mass shift introduced by the SILAC amino acids resulted in triplet peaks (i.e., the same peptide from three different time points) with the relative intensities equal to the relative abundance of the peptide. This SILAC approach allows for high-accuracy quantification of phosphopeptides with in most cases localization of the phosphate group with single amino acid accuracy. Two biological replicates were used to perform independent experiments for each ligand stimulation. (B) Labeling efficiency example of a peptide containing a lysine and arginine residue. The arrows indicate the position of partially labeled peptides.
