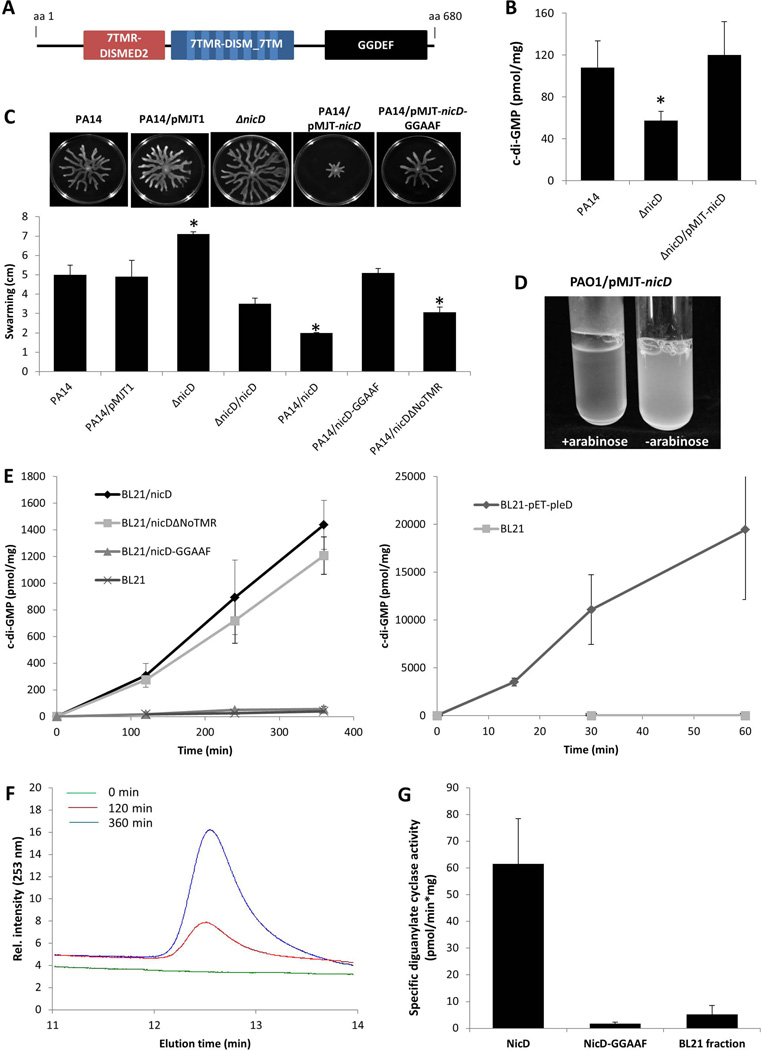
Figure 3. nicD harbors in addition to 7TMR-DISMED2 and 7TMR-DISM_7TM domains a GGDEF domain, and encodes an active diguanylate cyclase.
(A) NicD (PA4929) is predicted to be a membrane-bound protein harboring three domains including a periplasmic 7TMR-DISMED2 sensory domain, a membrane-spanning 7TMR-DISM_7TM domain and a cytoplasmic GGDEF domain. Model of NicD domain organization and putative subcellular localization of NicD domains. NicD comprises of three domains: an extracellular 7TMR-DISMED2 (7 Trans-membrane Receptor with Diverse Intracellular Signaling Modules extracellular) domain, a membrane-spanning 7TMR-DISM_7TM (7TMR-DISM, 7 Trans-membranes) domain, and a cytoplasmic diguanylate cyclase (GGDEF) domain. Domain localization is based on TMHMM prediction. (B) C-di-GMP levels in P. aeruginosa PA14, ΔnicD and ΔnicD/pMJT-nicD biofilms. C-di-GMP was quantitated using an HLC-based method with commercially available c-di-GMP as standard. c-di-GMP (pmol/mg) refers to c-di-GMP levels (pmol) per total cell pellet protein (in mg). Experiments were repeated at least three times. Error bars indicate standard deviation. *, significantly different from wild type biofilms; p < 0.05 as determined by ANOVA and SigmaStat (C) Swarming motility of P. aeruginosa PA14, ΔnicD, and strains overexpressing nicD, nicD-GGAAF, and nicDΔNoTMR. The swarming diameter was recorded following 48 hr of growth on NA agar. Representative images of swarming motility P. aeruginosa PA14, ΔnicD, and strains overexpressing nicD, nicD-GGAAF, and nicDΔNoTMR are shown above the graph. P. aeruginosa PA14 harboring the empty vector pMJT1 was used as vector control. Experiments were repeated at least three times. Error bars indicate standard deviation. *, significantly different from wild type biofilms; p < 0.05 as determined by ANOVA and SigmaStat. (D) Image of P. aeruginosa PAO1/pMJT-nicD planktonic culture prior to and following addition of arabinose to induce nicD expression. –arabinose, no arabinose was added; +arabinose, 1% arabinose was added to the growth medium. (E) Formation of c-di-GMP by total cell extracts of E. coli BL21 overexpressing nicD, nicD variants and pleD. DGC assays were carried out using a total of 200 µg of E. coli BL21 cell extracts overexpressing nicD and nicDΔNoTMR lacking the N-terminal 7TMR-DISMED2 domain (NicDΔNoTMR) or nicD-GGAAF in which the GGDEF motif was substituted with GGAAF under the control of the IPTG-inducible promoter of the pET101D vector or a total of 20 µg of E. coli BL21 cell extracts overexpressing pleD cloned into pET11b (Merritt et al., 2007, Paul et al., 2004) were used. Cell extracts of E. coli BL21 not harboring any vector (200 and 20 µg) were used as controls while E. coli BL21 overexpressing pleD were used as positive control. Experiments were carried out in triplicate. Error bars indicate standard deviation. (F) Elution profiles of c-di-GMP produced by purified NicD. The reaction mixtures were analyzed by HPLC for the presence of c-di-GMP 0, 120, and 360 min post initiation of DGC assays. Representative peaks corresponding to c-di-GMP are shown. (G). Specific diguanylate cyclase activity of purified NicD and NicD-GGAAF. Diguanylate cyclase assays were performed using 25 µM GTP and 70 µg of Ni-affinity chromatography purified protein. BL21 control, protein purified from E. coli cell extracts not harboring any vector was used as negative control. Experiments were carried out in triplicate. Error bars indicate standard deviation.