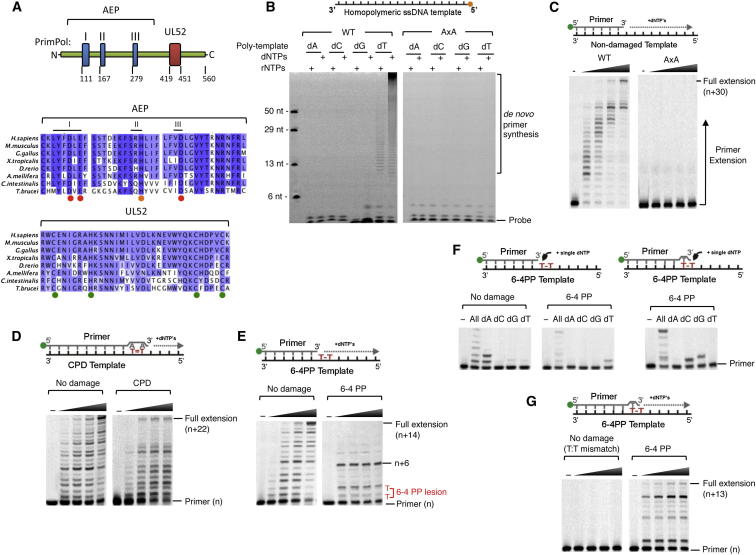
Figure 1.
Domain Architecture and Catalytic Activities of Human PrimPol
(A) Schematic and multiple sequence alignment of PrimPol conserved domains. The catalytic AEP domain containing three signature motifs (I, II, and III) and the UL52-like zinc finger domain are indicated, including amino acid number. Multiple sequence alignment was generated with a selection of PrimPol homologs; blue shading indicates ≥ 40% sequence identity, red circles indicate residues required for metal ion binding, orange circle for nucleotide binding, and green circles for chelation of zinc.
(B) Primer synthesis by wild-type (WT) His-tagged human PrimPol and catalytic mutant (AxA). Homopolymer DNA templates (500 nM) were incubated with dNTPs or rNTPs (500 μM), magnesium ions, and WT or AxA PrimPol (1 μM) for 2 hr at 37°C.
(C) DNA synthesis by PrimPol. Primer-template substrate (20 nM) and dNTPs (200 μM) were incubated with or without (−) PrimPol (WT or AxA; 50 nM) at 37°C for increasing times (2, 5, 10, 15 min).
(D–G) DNA synthesis by PrimPol on templates containing either a T-T cis-syn cyclobutane pyrimidine dimer (CPD) (D) or a T-T pyrimidine (6-4) pyrimidone photoproduct (6-4 PP) (E–G) was compared to PrimPol DNA synthesis on undamaged templates using primer extension assays as described in (C). CPD is annealed opposite two 3′ terminal dA residues, thereby testing PrimPol extension opposite the lesion (D). 6-4 PP is at bases +1 and +2 of template relative to 3′ terminus of primer to test read-through (E) and in the presence of single dNTPs for a single 30 min reaction to test nucleotide incorporation opposite 3′ T (F, middle panel). Primer with 3′ terminal dT opposite 3′ T of 6-4 PP was used to test nucleotide incorporation opposite 5′ T of lesion (F, right panel) and, when all dNTPs included, extension (G). Note: undamaged template in (G) contains a 3′ terminal T:T mismatch.