Figure 1.
Identification of Conserved POUV Targets
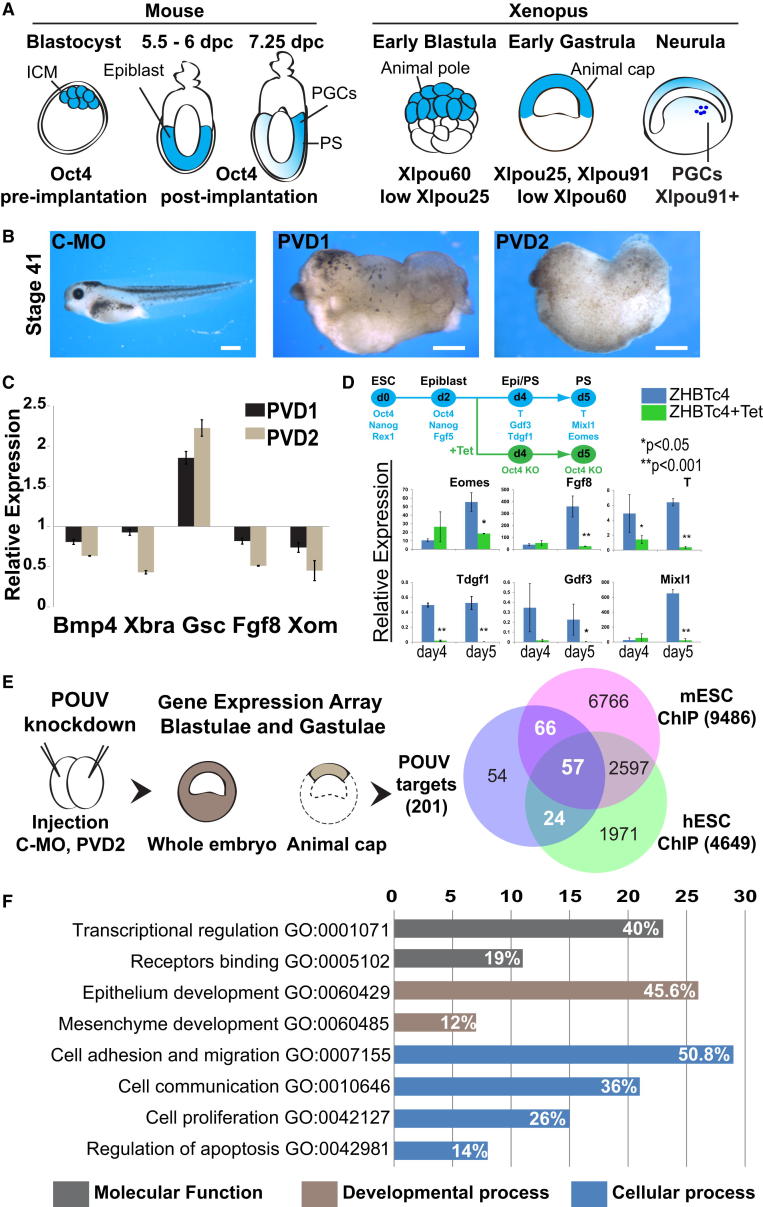
(A) Graphical representation of Oct4/POUV expression patterns in M. musculus and X. laevis.
(B) POUV morphant phenotypes. Two-cell-stage embryos were injected with 120 ng of Control-MO or morpholino combinations; POUV-depleted (PVD), a mixture of Xlpou25, Xlpou60, and Xlpou91 MOs; and POUV-depleted-2 (PVD2), the same mixture but with a new Xlpou25 MO targeting both pseudoalleles of Xlpou25. Tadpoles were photographed at stage 41. The scale bar represents 0.5 mm.
(C) qRT-PCR analysis of stage 10 embryos. Expression levels were normalized to Odc (ornithine decarboxylase) and are relative to the Control-MO. Data are shown as mean ± SD.
(D) ZHBTc4 mESCs expressing Oct4 as a tetracycline (Tet)-suppressible transgene were differentiated to mesendoderm. Tet was added as indicated and qRT-PCR performed to monitor gene expression in differentiation. Expression is normalized to TBP and relative to ESCs. Data are shown as mean ± SD. p values were calculated by ANOVA test of minimum of four independent experiments.
(E) POUV-dependent gene expression. Ectoderm (animal caps) from Control-MO and PVD2 embryos was dissected at early blastula stage (stage 8) and cultured until intact sibling embryos reached late blastulae (stage 9) and early gastrulae (stage 10) stages, as depicted. RNA from these explants was applied to Agilent 44K microarrays. POUV-regulated genes (N = 307) were identified by ANOVA analysis. Mammalian homologs (N = 201) of the POUV targets were identified by PSI-BLAST with minimum 60% identity. The Venn diagram shows the overlap of the genes regulated by POUV in Xenopus (N = 201) with global ChIP assays in murine and human ESCs.
(F) Enriched Gene Ontologies of the targets conserved in mouse and human (N = 57) show the number of genes included in each GO term. White values on the bars show the percentage of genes included in the conserved targets list (N = 57).