Abstract
The widespread paradigm in ecology that community structure determines function has recently been challenged by the high complexity of microbial communities. Here, we investigate the patterns of and connections between microbial community structure and microbially-mediated ecological function across different forest management practices and temporal changes in leaf litter across beech forest ecosystems in Central Europe. Our results clearly indicate distinct pattern of microbial community structure in response to forest management and time. However, those patterns were not reflected when potential enzymatic activities of microbes were measured. We postulate that in our forest ecosystems, a disconnect between microbial community structure and function may be present due to differences between the drivers of microbial growth and those of microbial function.
A major portion of Europe's forest cover (approx. 57% if the Russian Federation is excluded) is managed for woody biomass production1. Forest system management practices (FMPs) can significantly alter soil physical, biological and chemical properties, as well as microclimatic conditions. These in turn can have profound effects for many ecosystem functions including productivity, carbon storage and biodiversity conservation2,3,4. However despite their importance as drivers for most ecosystem functions, the influence of different forest management practices on microbial communities in different soil compartments and their functional traits is still not well understood. It is generally believed that the structure of microbial communities determines their function5,6,7,8,9. However, they also propose that there may be, in some instances, a disconnection between microbial community structure and its functions. For instance, findings from several ecosystems have demonstrated different responses between microbial community structure and functions9,10. Thus it can be difficult to predict how changes in environmental factors will affect microbial community structure and microbially-mediated ecosystem functions in different ecosystems because there may be differences in the resistance to change at either the the community and functional levels, and because of functional redundancy within the microbial community11. The aims of this study were to investigate the patterns of and connections between microbial community structure and microbially-mediated functions across different FMPs as a function of time.
Leaf litter degradation is a very interesting case study for examining microbial structure-function relationships. It provides an important basis for the nutrient supply of trees in most forest ecosystems and thus leaf litter degradation and recycling, which is mainly catalyzed by microbes, determines productivity to a large degree12. Forest system management practices can potentially change the chemical composition of leaf litter by changing the dominant tree species as well as tree species richness, abundance and performance2,12. In addition, microbial communities colonizing leaf litter undergo major temporal changes, both seasonally and over the course of litter decomposition13,14. In temperate forests, the bulk of the litter input (for both leaf and fine root litter) occurs in the autumn and the concentrations of easily degradable substrates from fresh leaf-litter including carbohydrates cellulose, proteins, starch and tannins are the highest in soil at this time. This presence of labile substrates may in turn stimulate microbial biomass in winter15,16,17,18. Plant and soil microbial seasonal cycles can also significantly influence nutrient availability in forest soil, also potentially affecting microbial community composition and feedbacks on microbial decomposition of leaf litter5. In this study we investigated microbial communities colonizing the litter material as well as selected functional traits over time in leaf litter from unmanaged stands and stands subject to different forest management regimes (different FMPs). The tested regimes involved age-class and selection cutting, both of which leave rather many forest gaps that enable the establishment of diverse tree species12,19. The unmanaged stands had fewer gaps and were only dominated by European beech (Fagus sylvatica), which produces low-quality leaf litter with a high C:N ratio12. Microbial community structure was determined broadly using phospholipid fatty acids. The functional traits of microbial communities were evaluated by measuring the potential activities of specific microbial enzymes involved in the biogeochemical cycling of important elements such as carbon, nitrogen and phosphorus10. We hypothesized that FMP and time of litter incubation in soil (days after incubation, DAI) both have significant effects on microbial community structure and potential enzyme activities. However the extent to which microbial community structure and function will be influenced will differ leading to a decoupling of structure–function relationships in this soil compartment. FMP and DAI may strongly affect microbial community structure, however, different microbial communities from different FMPs and/or DAI may perform similar enzyme activities (functional redundancy)10. Furthermore, differences between the drivers of microbial growth and enzyme activities may also be responsible for the disconnection of microbial community structure and function20. We used multivariate analysis to identify leaf litter properties with significant effects on the overall microbial community and enzyme activity patterns. Factors considered in this analysis included leaf litter quality parameters (C:N and lignin:N ratios) and other abiotic properties of the litter such as its pH, water content, and concentrations of microbial macronutrients and micronutrients (trace elements).
Results
Effects of FMP and DAI on microbial community structure and potential enzyme activities
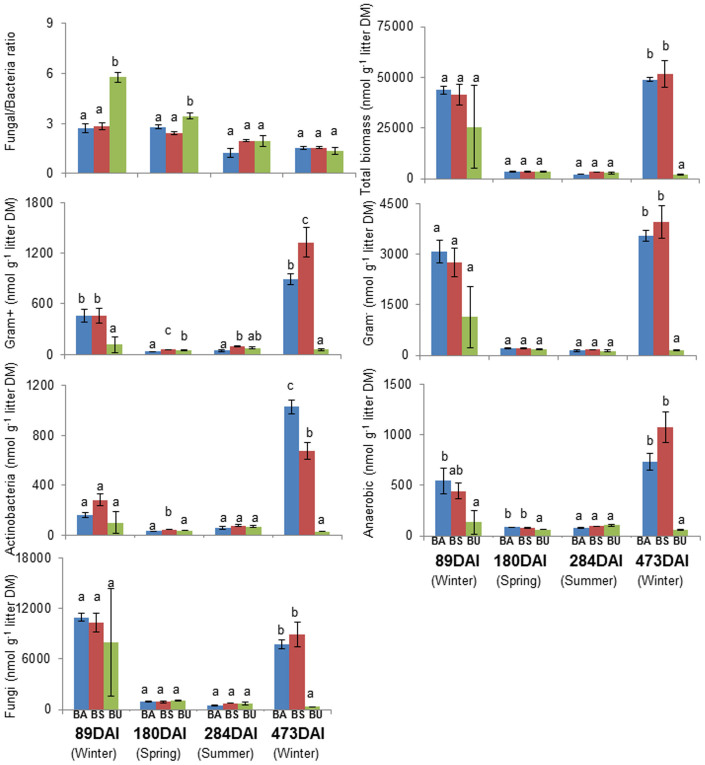
Abundance of phospholipid fatty acid (PLFA) indicators were significantly influenced by FMP and temporal (DAI) changes, and also by the interaction between FMP and DAI (P < 0.001 in all cases, Table S1, Figure 1). Overall in the leaf litter of managed forests (beech age-class, BA and beech selection cutting, BS) the highest amounts of PLFA (both total and key groups) were detected in winter (89 and 473 DAIs) while the lowest PLFA amounts were measured in spring and summer (180, 284 DAIs) (Figure 1). Similar results were obtained for litter material from the unmanaged beech forest (BU). However, during the second winter the microbial biomass in the BU forest did not increase to the same extent as in the BA and BS forests. The fungal: bacterial ratio decreased over time under all FMPs (Figure 1). At 89 and 180 DAI, the leaf litter from BU forest had a significantly higher fungi: bacteria ratio than that observed in the BA and BS forests (89 DAI: BU = 5.76, BS = 2.82, BA = 2.72; 180 DAI: BU = 3.43, BS = 2.42, BA = 2.78). PLFA indicators for all bacterial groups except Gram negative bacteria varied significantly with FMP in spring. In summer, no significant differences among different PLFA indicators were ascertained for bacterial groups, except for Gram positive bacteria whose indicators were highest in BS forest and lowest in BA forest. NMDS analysis of PLFA patterns revealed four distinct clusters representing samples taken at different DAI (RDAI = 0.97, P < 0.001, Figure 2a). The different FMPs also affected microbial community structure, as shown by the existence of one cluster corresponding to samples from managed forests (BA and BS) and another corresponding to the BU forest at specific DAI values (RFMP = 0.69, P < 0.001, Figure 2a).
Figure 1. Fungal: bacterial ratio and microbial PLFA abundances (both total and key groups) (mean ± SE) in leaf litter samples collected at four sampling dates (days after incubation commenced, DAI).
European beech age-class forest (BA, blue), European beech selection cutting forest (BS, red) and unmanaged deciduous forest reserves dominated by European beech (BU, green). Litter DM = litter dry mass. Different letters indicate significant differences (P < 0.05) between different treatments at each DAI according to one-way ANOVA and Fisher's Least Significant Difference (from 89 DAI to 473 DAI).
Figure 2. Nonmetric multidimensional scaling (NMDS) ordination of (a) microbial community structure inferred from PLFA analysis and (b) activities of 8 different enzymes.
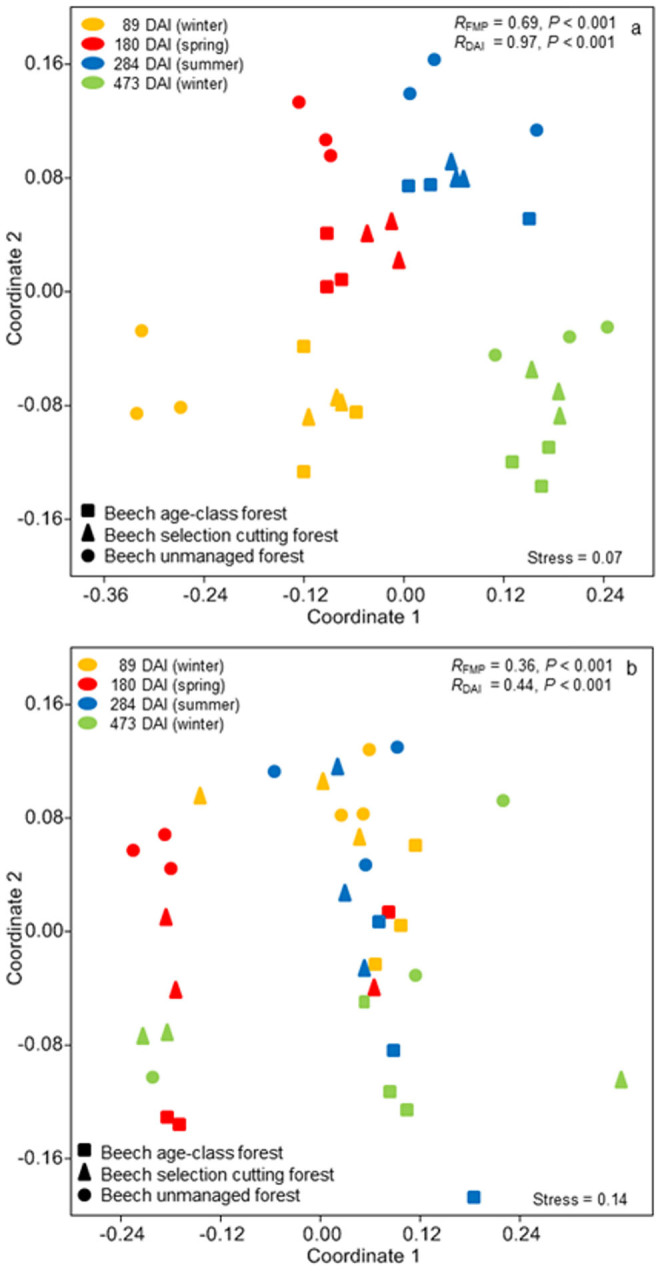
FMP = forest system management practice; DAI = days after incubation commenced; R = degree of separation between test groups ranging from −1 to 1; R = 0, not different; R = 1, completely different; P values were based on 999 permutations.
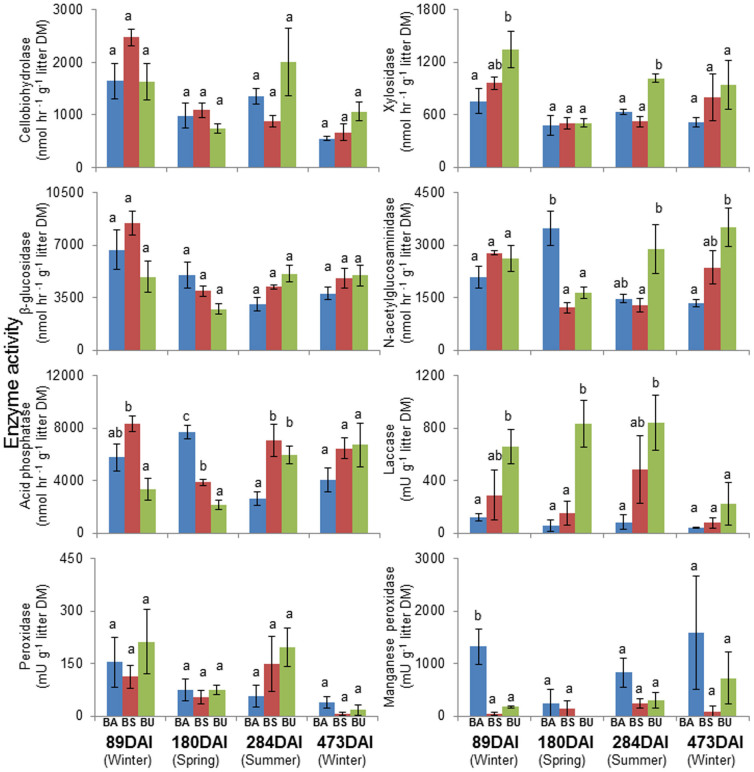
The potential activities of the enzymes investigated in this study did not follow the same trends as lipid biomass or lipid indicators (Tables S1 and S2, Figures 2b and 3). Mantel tests revealed an insignificant correlation between the PLFA and enzyme activity matrices (Mantel R (Euclidean distance matrix) = 0.05, P > 0.05, Mantel R (Bray-Curtis distance matrix) = 0.09, P > 0.05). Enzyme activities did not vary according to the microbial biomass (Table S2). Enzyme activities in leaf litter under different forest management practices are presented in detail in Figure 3. Hydrolytic enzymes, cellobiohydrolase and β-glucosidase were significantly influenced by DAI and the interaction between FMP and DAI (P < 0.05, Table S1) but were not significantly influenced by FMPs. Xylosidase activity was significantly influenced by FMP and DAI (P < 0.01, Table S1). Xylosidase activity was generally high in BU (low forest management intensity) compared with BA (high forest management intensity). N-acetylglucosaminidase and acid phosphatase activities were significantly influenced by FMP and the interaction between FMP and DAI (P < 0.05, Table S1). The activities of the studied oxidative enzymes also behaved differently compared to microbial biomass: laccase activity was significantly affected by FMP and DAI (P < 0.05, Table S1), peroxidase by DAI (P < 0.01, Table S1), and manganese peroxidase by FMP (P < 0.01, Table S1). However, the interaction between FMP and DAI did not significantly affect the activities of any studied oxidative enzyme (P > 0.05, Table S1). Overall the observed NMDS patterns for microbial community structure were not reproduced in the NMDS analysis of enzyme activities where there was no recognizable clustering of samples representing either different FMPs or DAI values (Figure 2b). Although the different FMPs and DAI significantly influenced potential enzyme activity patterns, the degree of influence indicated by ANOSIM R values were much lower than for the microbial lipid abundances (RFMP = 0.36, RDAI = 0.44, P < 0.001 in both cases). In some cases, microbial communities collected from forests subject to different FMPs or collected at different times exhibited similar enzyme activities (Figure 2b).
Figure 3. Enzyme activities (mean ± SE) in leaf litter samples collected at four sampling dates (days after incubation commenced, DAI).
European beech age-class forest (BA, blue), European beech selection cutting forest (BS, red) and unmanaged deciduous forest reserves dominated by European beech (BU, green). Litter DM = litter dry mass. Data on oxidative enzyme activities are derived from Purahong et al.12. Different letters indicate significant differences (P < 0.05) between different treatments at each DAI according to one-way ANOVA and Fisher's Least Significant Difference (from 89 DAI to 473 DAI).
The effects of the physicochemical properties of leaf litter on microbial community structure and function
Our results indicated that microbial community structure and potential enzymatic activities are regulated and controlled by different physicochemical properties of the leaf litter. The structure of the microbial communities was significantly influenced by all leaf litter parameters (i.e. C:N and lignin:N ratios), the levels of most microbial macronutrients other than P (C, N, Mg, K, Ca, Fe), the levels of one microbial micronutrient (Mn), and the pH (Table 1). In contrast, only C, K, P, Mn, and pH significantly influenced enzyme activities. Leaf litter quality parameters (C:N and lignin:N ratios) that had very significant effects on the structure of the microbial community (P < 0.01) did not significantly affect the pattern of enzymatic activities (P > 0.05) (Table 1). Changes in leaf litter chemistry over time are presented in Figure S1.
Table 1. Goodness-of-fit statistics (R2) of environmental factors fitted to the nonmetric multidimensional scaling (NMDS) ordination of microbial community structure and enzyme activities. The significance was based on 999 permutations. Significant factors (P < 0.05) are indicated in bold: *P < 0.05, ** P < 0.01, ***P < 0.001.
| Microbial biomass | Enzyme activities | |||
|---|---|---|---|---|
| Varibles | R2 | P | R2 | P |
| Total C | 0.701 | 0.001*** | 0.266 | 0.007** |
| Total N | 0.424 | 0.001*** | 0.073 | 0.309 |
| C:N ratio | 0.717 | 0.001*** | 0.032 | 0.566 |
| Lignin:N ratio | 0.280 | 0.006** | 0.090 | 0.209 |
| pH (CaCl2) | 0.460 | 0.001*** | 0.315 | 0.002** |
| pH (H2O) | 0.086 | 0.258 | 0.025 | 0.634 |
| Mg | 0.235 | 0.009** | 0.010 | 0.872 |
| K | 0.396 | 0.001*** | 0.608 | 0.001*** |
| Ca | 0.288 | 0.006** | 0.063 | 0.354 |
| P | 0.052 | 0.414 | 0.352 | 0.002** |
| Mn | 0.271 | 0.010** | 0.218 | 0.013* |
| Fe | 0.383 | 0.001*** | 0.000 | 0.999 |
| Cu | 0.006 | 0.910 | 0.026 | 0.650 |
| Co | 0.073 | 0.265 | 0.009 | 0.919 |
| V | 0.001 | 0.988 | 0.119 | 0.149 |
| Water content | 0.103 | 0.166 | 0.085 | 0.216 |
| Total lignin | 0.094 | 0.211 | 0.086 | 0.218 |
Discussion
In this experiment, we examined the patterns of and connections between microbial community structure and function across different forest management practices and as a function of time using leaf litter as an example. PLFA analysis was used for characterizing the microbial community structure. This method provides a broad estimation of the viable biomass of different microbial functional groups21. Some phospholipids (such as the fungal indicators 18:1ω9, 18:2ω6,9 and 18:3ω3,6,9) or potential enzymatic activities could be derived from plants22. While this consideration was taken into account for our interpretations, we assume that residual contributions from plants would be far surpassed by new microbial growth. This assumption is based on sample timing at 89 DAI to 473 DAI. By 89 DAI, a residual effect from PLFA or enzymes from the residual litter would be minimal, especially because the litter was dried and processed before plot establishment23. Based on the results of leaf litter decomposition rates of the same experiment, we showed that forest management practices significantly influence the decomposition kinetics (BA forest has higher decomposition rate than BU forest)12. Thus, a caveat to this dataset is that at the same DAIs, leaf litter from different FMPs (BA and BU forests) may not be at the same decomposition stages. This may also affect the patterns of enzyme activities and PLFA in leaf litter at the same DAI (different FMPs).
As expected, high microbial biomass was observed for all FMPs in winter (89 and/or 473 DAI)15,16,17,18. Our biomass analyses indicated that fungi were the dominant leaf litter-decomposing organisms under all of the studied FMPs and DAI. On the other hand, bacteria increased in abundance as decomposition progressed. Fungi are generally considered to be the primary decomposers of leaf litter in temperate forest ecosystems13 and they rapidly colonize leaf litter during the early stages of decomposition24. Biological fragmentation caused by the action of fungi and invertebrates and physical fragmentation results in an increased leaf litter surface area and the release of nutrients during the later stages of leaf litter decomposition25, which may enhance the activity of bacteria during litter decomposition26.
We identified clear differences between the microbial communities from leaf litter produced under different FMPs (i.e. from managed vs. unmanaged forests) and between microbial communities collected at different time points within the 473 day sampling period. This strongly suggests that the structure of microbial communities in the leaf litter of the studied forest systems is sensitive to forest management practices, the stage of litter decomposition, and seasonal factors. Similar conclusions have been drawn in a number of previous studies examining various types of disturbance: in general, the structures of microbial communities are sensitive to many factors and not immediately resilient to disturbance11. In our studied sites, FMPs affect tree species composition that, in turn, significantly alters leaf litter quality and nutrients in BA and BS forests as compared to BU forest12. The results from multivariate analysis (Table 1) demonstrate that leaf litter quality and most macronutrients are significantly correlated with changes in microbial community composition. BU forest is located in a forest reserve area, which, for at least 60 years, has not undergone thinning, wood harvesting, or other serious disturbances that would have resulted in the loss of large numbers of trees. Thus the BU had fewer gaps and were only dominated by European beech (Fagus sylvatica), which produces low-quality leaf litter12. On the other hand, thinning operation and woody biomass harvesting in BA (whole stand harvesting) and BS (individual tree harvesting) could generate the forest gaps that are important for natural tree regeneration and the maintenance of plant diversity. Thus, Acer sp. successfully established by natural regeneration in both BA and BS forests (Table S3). In addition, a small fraction of Fraxinus sp. was also found in BA forest (Table S3). Acer sp. and Fraxinus sp. leaf litter are known to have better quality and nutrients compared with Fagus sylvatica leaf litter12.
In contrast, we did not identify any clear changes in the patterns of potential enzyme activities in leaf litter samples due to FMPs or temporal factors. The responses of enzymatic activities to DAI and FMPs varied and were not always significant. Our results therefore indicate a decoupling of microbial community structure and the metabolic functions of potential microbial enzymatic activities across FMPs and temporal changes in temperate forest ecosystems. This might be explained, at least in part, by the widespread occurrence of hydrolytic enzymes (including polymer-disintegrating cellulases and xylanases) among different groups of microbes (fungi, bacteria, protists) so that they can replace each other in the course of community changes (functional redundancy)27. For example, the cellulases of fungi may disappear along with their producers during leaf litter succession, but they can be replaced by similarly active enzymes of actinobacteria or other Gram-positive bacteria28. In the case of oxidative enzymes, the situation is somewhat different. Oxidative enzymes were reported to be more organism-specific. Both laccase and generic peroxidases are primarily secreted by fungi (both ascomycetes and basidiomycetes)29,30. This is especially true for Mn-peroxidase that, in soil environments, is exclusively produced by specific basidiomycetous fungi causing a kind of white-rot in leaf litter31,32. Thus, the activities of oxidative enzymes may not be as widely spread as those of hydrolytic enzymes. This means, if appropriate fungi are present in a litter bag, there will be a high level of oxidative enzymes but if not, this activity will be lacking.
Insofar as our results deviate from the generally notion that structure determines function, these findings together with those of Frossard et al. can be regarded as supporting a paradigm shift10. Frossard and colleagues demonstrated a decoupling of bacterial community structure and function (represented by the activities of 10 separate enzymes) across temporal changes in nascent stream corridors. The disconnection between microbial community structure and function in these two cases may be due to functional redundancy within the studied microbial communities10. Our results show that different microbial communities (from either different FMPs or different points in time) can produce similar enzyme activities, thereby we speculate that at least some level of functional redundancy may occur. In addition to microbial functional redundancy, we demonstrated that differences in the drivers of microbial growth versus function can be used to explain the uncoupling between microbial community structure and the metabolic activity of microbial enzymes20. While all of the studied litter quality parameters and the levels of microbial macronutrients (except P) significantly influenced microbial community structure, only a few of these factors were found to significantly affect the overall pattern of potential enzyme activities. Vice versa P concentration did not significantly influence the microbial community structure but did have significant effects on the overall patterns of enzyme activity.
Based on our results, we suggest that there is some degree of functional redundancy in the microbial community. However, we also detected differences in litter decomposition rates and nutrient dynamics between forests with different FMPs (especially between unmanaged and age-class forests)12. This is due to the time lag in functional similarity. Our results show that microbial communities from different FMPs perform similar enzyme activities at different points in time. We have previously shown that the rates of leaf litter decomposition are highest in systems that decompose lignin most rapidly (i.e. those with high levels of manganese peroxidase, an efficient ligninolytic enzyme)12. Thus, leaf litter produced under FMPs that are more amenable to rapid colonization by microbial communities producing efficient ligninolytic enzymes may decompose relatively quickly.
In summary, the structure of microbial communities and microbial ecological functions of metabolic enzymes are dependent on the complex interactions between numerous environmental factors. In temperate forest ecosystems, forest management is one of the most important factors affecting both microbial community structure and its ecological function.
Methods
Study site
The study was conducted at three forest sites in the Hainich-Dün region of Central Germany (about 1,300 km2; 51°16′N 10°47′E), which is part of the German Biodiversity Exploratories33. The forests in this region grow on soils over limestone bedrock33. The main soil type in the study area according to the German soil classification system and the World Reference Base of Soil Resources is a Stagnosol33. The soil pH is weakly acidic (5.1 ± 1.1; mean ± SD)34. Thin litter (in the range 2–5 cm) were recorded at all three forest sites12. The annual mean temperature and precipitation ranged, respectively, from 6.5–8°C and 500–800 mm33. All information pertaining to these forests has been described in detail elsewhere12,33.
Three forest sites (10,000 m2 each) were selected based on their forest system management practices with each site representing one treatment. All sites were dominated by European beech (Fagus sylvatica) and located less than 30 km apart, to reduce the influence of variations in geography and climate. Within each forest site, we assigned three plots (2 × 8 m2) located on flat land to represent three experimental replicates of each forest management practice. The three FMPs considered in this study were: (i) European beech age-class forest (BA, semi-natural forest with natural regeneration, even-age forest structure); (ii) European beech selection cutting forest (BS, near-to-nature forest management with natural regeneration, uneven-age forest structure); and (iii) unmanaged deciduous forest reserves dominated by European beech (BU, uneven-age forest structure)12. These three forest sites differ in terms of the silvicultural management intensity indicator (SMI)35. The SMI was low for the BU and BS forests but high in the BA forest35. The litter composition for individual forest sites and the initial chemical composition of the litter material are given in Tables S3 and S4.
Experimental design and sampling
At each forest site, freshly fallen leaves of deciduous species were collected from the forest floor in October, 2009. The leaves from each plot were separated according to species and air dried to constant weight at room temperature. Ten grams of local mixed leaves, representative of the litter composition of the deciduous tree species at each respective site were then placed in litterbags (25 × 25 cm, 2 mm nylon mesh size) for deployment12. At the end of the litter fall period (13 November 2009), 12 replicate litter bags were placed in each plot, for a total of 36 litterbags per management type and 108 litterbags in total. 27 additional litterbags (9 per treatment) were retained to determine the initial dry mass (oven-dried at 105°C ≥ 24 h until constant weight), nutrient element concentrations, and lignin content of the litter mixtures (Data supplement, Table S4). To preserve the situation in the temperate forest, we allowed the fresh leaf canopy litter to fall above the litter bags. Litterbags were retrieved on four sampling dates: in 2010 on 10 February (89 days after incubation commenced, DAI), 12 May (180 DAI), 24 August (284 DAI), and in 2011 on 1 March (473 DAI). On each sampling occasion, 3 replicate litter bags per plot (nine litterbags per treatment) were randomly removed, placed into a separate clean plastic bag to reduce the loss of small fragments, and transported on ice (0°C) to the laboratory within 4 h. Once in the laboratory, litter bags were processed immediately. First, the three replicate litterbags retrieved from the same plot and treatment were pooled and their wet weight was determined. Thus, three composite samples from each site were obtained for each treatment and DAI. Each composite sample was then homogenized and subsampled for determination of dry mass, nutrient element concentrations, lignin content, phospholipid fatty acids analysis (PLFA) and enzyme activities.
Details of phospholipid fatty acid analysis (PLFA)
Microbial community structure was assessed using the PLFA method described by Wu et al.36. Briefly, phospholipids were first extracted three times from leaf litter using a methanol, chloroform, citric acid solution. Phospholipids were then separated using silica columns and methylated by alkaline methanolysis. A total of 42 lipids with 20 or fewer carbon atoms each were used to calculate the total microbial lipid abundance (nmol lipid g litter DM−1). Several individual PLFAs were used as indicators for key groups of microbes: 15:0 iso for Gram-positive bacteria; 16:1ω7c for Gram-negative bacteria; 17:0 cyclo and 19:0 cyclo for anaerobic bacteria; 10Me16:0 for Actinobacteria and 18:2ω6,9c for general fungi. In our study, 3–5 individual PLFA markers that can be used to quantify the abundances of Gram-positive bacteria (15:0 iso, 15:0 ante, 16:0 iso, 17:0 iso and 17:0 ante), Gram-negative bacteria (16:1ω5c, 16:1ω7c and 16:1ωw9c) and general fungi (18:1ω9, 18:2ω6,9c and 18:3ω3,6,9c) were highly correlated (R = 0.93–1.00, P < 0.001; Table S5)5,37,38,39. Thus, we found that results were similar whether we used one versus more indicators for each microbial group. For our final analysis, we only used one indicator (except for anaerobic bacteria).
Determination of potential enzymatic activities
A total of 8 enzyme activities were measured. Specifically, five hydrolytic enzymes which are important for carbon (β-glucosidase (EC 3.2.1.21), cellobiohydrolase (EC 3.2.1.91), and xylosidase (EC 3.2.1.37), nitrogen (N-acetylglucosaminidase, EC 3.1.6.1), and phosphorus (acid phosphatase, EC 3.1.3.2) acquisition were assessed based on methods described by Sinsabaugh et al. and German et al.40,41. Three oxidative enzymes important for lignin degradation (laccase (EC 1.10.3.2), general peroxidase (EC 1.11.1.7), and manganese peroxidase (EC 1.11.1.13), MnP) were measured as described by Purahong et al.12. Manganese peroxidase activities were not captured by the assay for general peroxidases because, without sufficient amounts of Mn(II) in the assay solution, manganese peroxidase cannot work. Thus in this experiment we found no correlation between the activities of these two enzymes (R = 0.09, P = 0.582) (Table S2).
Measurements of physicochemical parameters of the leaf litter
The water content and pH (in H2O and 0.01 M CaCl2) of all leaf litter samples were also measured. The pH measurement in 0.01 M CaCl2 was found to provide more consistent and reproducible results than pH measured in H2O. Total C and N were determined by dry combustion at 1,000°C with an Elementar Vario EL III elemental analyzer (Elementar Analysensysteme GmbH, Hanau, Germany) (DIN/ISO 10694 (Aug. 1996)). Nutrient ions (Mg, K, P, Ca, Fe, Cu, V, Mn, Co) were determined using inductively coupled plasma (ICP) optical emission spectrometry (ICP-OES) and mass spectrometry (ICP-MS) according to manufacturers' specifications. Total lignin was calculated by summing Klason lignin (acid insoluble lignin) and acid soluble lignin42. Klason lignin content was determined gravimetrically as the dry mass of solids after sequential hydrolysis with sulfuric acid (72% w/w); in a second step, acid soluble lignin was measured UV-photometrically in 4% H2SO443,44. All physicochemical analyses were conducted in triplicate on the same subsample.
Statistical analysis
Total microbial biomass, the abundance of microbial indicators, and enzyme activities were analyzed using two-way (FMP and DAI) analysis of variance (ANOVA) with P < 0.05 considered significant, implemented in the PAST program12,45. All data were also analyzed using a one-way ANOVA to determine the differences (P < 0.05) among different treatments at each DAI12. When the differences were significant, they were further analyzed using a post-hoc test (Fisher's Least Significant Difference, P < 0.05). All datasets were tested for normality using Shapiro-Wilk and Jarque-Bera tests and the equality of group variances was examined using Levene's test. ANOVA residuals were also plotted to determine the normal probability using the Shapiro-Wilk W test. A Log10 transformation (log10 (x + 1)) was applied to all data sets that did not meet the parametric assumptions. Non-metric multidimensional scaling (NMDS) based on a Bray-Curtis similarity measures (100 random restarts) were performed on a matrix of PLFAs information (expressed as mole percentages) and a matrix including information on all enzyme activities (Log10 transformation) using PAST program45 and vegan package of R version 2.13.146. Two-way (FMP and DAI) Analysis of Similarity (ANOSIM) based on Bray-Curtis measure was conducted to test for differences among clusters45. Correlation between microbial community structure (PLFA matrix) and function (enzyme activity matrix) were analyzed using Mantel tests in PAST program45. To investigate the influence of leaf litter physicochemical factors on patterns of microbial community and enzyme activities, we fitted the physicochemical factors onto NMDS ordinations and calculated goodness-of-fit statistics (R2) using the envfit function in the vegan package46. P values were based on 999 permutations46.
Author Contributions
D. Krüger, F.B., M.H., J.G., D. Kapturska, M.P. and W.P. conceived and designed the experiments. M.P., D. Kapturska, W.P., V.D. and S.M. performed the experiments. W.P. analyzed the data. D.Krüger, F.B., M.H., J.G. and W.P. contributed reagents/materials/analysis tools. W.P., M.S., M.P., D. Krüger, M.H., J.G. wrote the paper.
Supplementary Material
Supplementary information
Acknowledgments
This work was funded in part by DFG Priority Program 1374 on ‘Infrastructure-Biodiversity-Exploratories’ (KR 3587/1-1, KR 3587/3-2, SCHL 446/13-2, HO 1961/4-1, HO 1961/5-2). W. Purahong and D. Kapturska also were kindly supported by the Helmholtz Impulse and Networking Fund via the Helmholtz Interdisciplinary Graduate School for Environmental Research (HIGRADE). The funders DFG and HIGRADE had no input into the study design, the data collection and analysis, the decision to publish, or the preparation of the manuscript. We thank the managers of the three exploratories, Swen Renner, Sonja Gockel, Kerstin Wiesner, and Martin Gorke for their work in maintaining the plot and project infrastructure; Simone Pfeiffer and Christiane Fischer for giving support through the central office, Michael Owonibi for managing the central data base, and Markus Fischer, Eduard Linsenmair, Dominik Hessenmöller, Jens Nieschulze, Daniel Prati, Ingo Schöning, Ernst-Detlef Schulze, Wolfgang W. Weisser and the late Elisabeth Kalko for their role in setting up the Biodiversity Exploratories project.
References
- FAO. [Productive functions of forest resources]. Global forest resources assessment 2010 main report [Rojas-Briales, E. (ed.)] [87–88] (Food and Agriculture Organization of the United Nations, Rome, 2010). [Google Scholar]
- Waldrop M. P., McColl J. G. & Powers R. F. Effects of forest postharvest management practices on enzyme activities in decomposing litter. Soil Sci. Soc. Am. J. 67, 1250–1256 (2003). [Google Scholar]
- Purahong W. et al. Changes within a single land-use category alter microbial diversity and community structure: Molecular evidence from wood-inhabiting fungi in forest ecosystems. J. Environ. Manage. 139, 109–119 (2014). [DOI] [PubMed] [Google Scholar]
- Purahong W. et al. Comparing fungal richness and community composition in coarse woody debris in Central European beech forests under three types of management. Mycol. Prog. 13, 959–964 (2014). [Google Scholar]
- Kaiser C. et al. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 187, 843–858 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire K. L. & Treseder K. K. Microbial communities and their relevance for ecosystem models: Decomposition as a case study. Soil Biol. Biochem. 42, 529–535 (2010). [Google Scholar]
- Talbot J. M. et al. Independent roles of ectomycorrhizal and saprotrophic communities in soil organic matter decomposition. Soil Biol. Biochem. 57, 282–291 (2013). [Google Scholar]
- Waldrop N. P. & Firestone M. K. Response of Microbial Community Composition and Function to Soil Climate Change. Microb. Ecol. 52, 716–724 (2006). [DOI] [PubMed] [Google Scholar]
- Gutknecht J., Goodman R. M. & Balser T. C. Linking soil process and microbial ecology in freshwater wetland ecosystems. Plant Soil 289, 17–34 (2006). [Google Scholar]
- Frossard A., Gerull L., Mutz M. & Gessner M. O. Disconnect of microbial structure and function: enzyme activities and bacterial communities in nascent stream corridors. ISME J. 6, 680–691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison S. D. & Martiny J. B. H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA. 105, 11512–11519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purahong W. et al. Influence of different forest system management practices on leaf litter decomposition rates, nutrient dynamics and the activity of ligninolytic enzymes: a case study from Central European forests. PLoS ONE 9, e93700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voříšková J. & Baldrian P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7, 477–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead D. L. & Sinsabaugh R. L. A theoretical model of litter decay and microbial interaction. Ecol. Monogr. 76, 151–174 (2006). [Google Scholar]
- German D. P. et al. Substrate concentration and enzyme allocation can affect rates of microbial decomposition. Ecology 92, 1471–1480 (2011). [DOI] [PubMed] [Google Scholar]
- Rinkes Z. L., Weintraub M. N., DeForest J. L. & Moorhead D. L. Microbial substrate preference and community dynamics during decomposition of Acer saccharum. Fungal Ecol. 4, 396–407 (2011). [Google Scholar]
- Moorhead D. L. & Sinsabaugh R. L. A theoretical model of litter decay and microbial interactions. Ecol. Monogr. 76, 151–174 (2006). [Google Scholar]
- Lipson D. A., Schmidt S. K. & Monson R. K. Carbon availability and temperature control the post-snowmelt decline in alpine soil microbial biomass. Soil Biol. Biochem. 32, 441–448 (2000). [Google Scholar]
- Schliemann S. A. & Bockheim J. G. Methods for studying treefall gaps: a review. Forest Ecol. Manag. 261, 1143–1151 (2011). [Google Scholar]
- Schimel J., Balser T. C. & Wallenstein M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394 (2007). [DOI] [PubMed] [Google Scholar]
- White D. C., Meadows P., Eglinton G. & Coleman M. L. In-situ measurement of microbial biomass, community structure and nutritional-status. Philos. Trans. R. Soc. Lond. Ser. A Math. Phys. Sci. 344, 59–67 (1993). [Google Scholar]
- Kaiser C., Frank A., Wild B., Koranda M. & Richter A. Negligible contribution from roots to soil-borne phospholipid fatty acid fungal biomarkers 18:2ω6,9 and 18:1ω9. Soil Biol. Biochem. 42, 1650–1652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esperschütz J. et al. Incorporation of carbon from decomposing litter of two pioneer plant species into microbial communities of the detritusphere. FEMS Microbiol. Lett. 320, 48–55 (2011). [DOI] [PubMed] [Google Scholar]
- Purahong W. & Hyde K. D. Effects of fungal endophytes on grass and non-grass litter decomposition rates. Fungal Divers. 47, 1–7 (2011). [Google Scholar]
- Word D. A. Using leaf litter breakdown to assess the effects of mountaintop removal mining on headwater streams in eastern Kentucky. (MS Thesis, University of Louisville, 1997). [Google Scholar]
- Plowman N. S. Impact of invertebrates and fungi on leaf litter decomposition across a forest modification gradient in Sabah, Malaysia. (MS Thesis, Imperial College London, 2012). [Google Scholar]
- Wilson D. B. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14, 259–263 (2011). [DOI] [PubMed] [Google Scholar]
- Wilson D. B. Biochemistry and genetics of actinomycete cellulases. Crit. Rev. Biotech. 12, 45–63 (1992). [DOI] [PubMed] [Google Scholar]
- Baldrian P. Fungal laccases - occurrence and properties. FEMS Microbiol. Rev. 30, 215–42 (2006). [DOI] [PubMed] [Google Scholar]
- Floudas D. et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719 (2012). [DOI] [PubMed] [Google Scholar]
- Steffen K. T., Hofrichter M. & Hatakka A. Mineralisation of 14C-labelled synthetic lignin and ligninolytic enzyme activities of litter-decomposing basidiomycetous fungi. Appl. Microbiol. Biotechnol. 54, 819–25 (2000). [DOI] [PubMed] [Google Scholar]
- Steffen K. T., Cajthaml T., Snajdr J. & Baldrian P. Differential degradation of oak (Quercus petraea) leaf litter by litter-decomposing basidiomycetes. Res. Microbiol. 158, 447–455 (2007). [DOI] [PubMed] [Google Scholar]
- Fischer M. et al. Implementing large-scale and long-term functional biodiversity research: The Biodiversity Exploratories. Basic Appl. Ecol. 11, 473–485 (2010). [Google Scholar]
- Naether A. et al. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 78, 7398–7406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall P. & Ammer C. How to quantify forest management intensity in Central European Forests. Eur. J. Forest Res. 132, 379–396 (2013). [Google Scholar]
- Wu Y. et al. Relationships between soil microorganisms, plant communities, and soil characteristics in Chinese subtropical forests. Ecosystems. 15, 624–636 (2012). [Google Scholar]
- Frostegard A. & Baath E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 22, 59–65 (1996). [Google Scholar]
- Frostegard A., Tunlid A. & Baath E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 43, 1621–1625 (2011). [Google Scholar]
- Kramer S., Marhan S., Haslwimmer H., Ruess L. & Kandeler E. Temporal variation in surface and subsoil abundance and function of the soil microbial community in an arable soil. Soil Biol. Biochem. 61, 76–85 (2013). [Google Scholar]
- Sinsabaugh R. L., Saiya-Cork K. & Long T. Soil microbial activity in a Liquidambar plantation unresponsive to CO2 driven increase in primary production. Appl. Soil Ecol. 24, 263–271 (2003). [Google Scholar]
- German D. P. et al. Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol. Biochem. 43, 1387–1397 (2011). [Google Scholar]
- Raiskila S. et al. FTIR spectroscopic prediction of Klason and acid soluble lignin variation in Norway spruce cutting clones. Silva Fenn. 41, 351–371 (2007). [Google Scholar]
- Effland M. J. Modified procedure to determine acidinsoluble lignin in wood and pulp. Tappi 60, 143–144 (1977). [Google Scholar]
- Liers C., Arnstadt T., Ullrich R. & Hofrichter M. Patterns of lignin degradation and oxidative enzyme secretion by different wood- and litter-colonizing basidiomycetes and ascomycetes grown on beech-wood. FEMS Microbiol. Ecol. 78, 91–102 (2011). [DOI] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T. & Ryan P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9 (2001). [Google Scholar]
- Oksanen J. Multivariate analysis of ecological communities in R: vegan tutorial. (2013). at <http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf> Date of access: 07/04/2014
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information