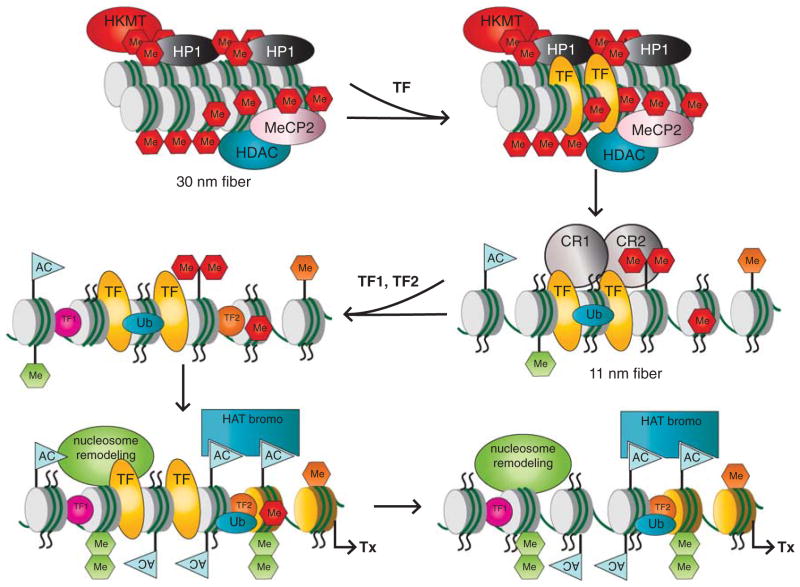
Figure 3.
A general model of gene activation of tissue-specific genes during embryonic eye development. A condensed chromatin domain, shown as 30 nm fiber, is marked by the presence of compacted nucleosomes marked by repressive PTMs (red octagons), and the presence of chromatin remodeling enzymes such as HDACs. The process of gene activation is initiated through binding of a TF (yellow) that can recognize its target sites in ‘closed’ chromatin conformation. Recruitment of distinct chromatin remodeling complexes/enzymes (for example, CR1 and CR2) through TF results in the generation of a novel set of histone PTMs, enriched for activating modifications (green octagons-methylations, blue flags-acetylations) and transition to the 11 nm chromatin conformation. Within this ‘open’ chromatin structure, TF1 (purple) and TF2 (orange), can locate their corresponding binding sites, and recruit additional nucleosome remodeling activities by reading the histone code that promotes transcription (Tx). It is possible that binding of the original TF (yellow) is no longer required at the binding site(s). See text for additional details.