Figure 4.
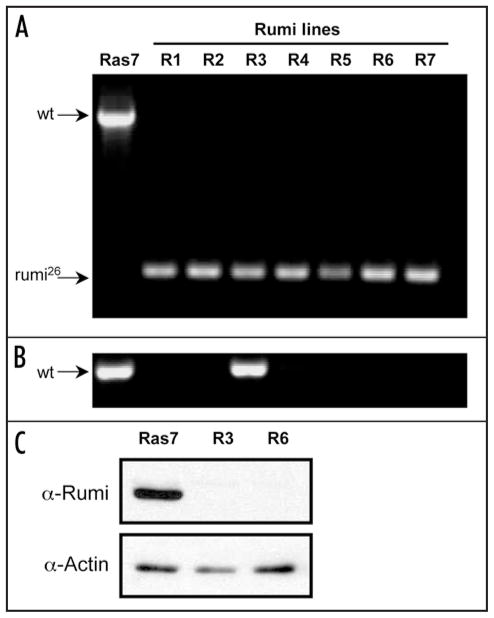
Cell lines are deficient for Rumi. Diagnostic PCR tests and Western blotting were used to genotype cells in the Rumi lines and determine their protein expression levels. Together the results show that 6/7 Rumi lines were pure populations of rumi26 homozygous cells. One line had a fraction of non-mutant cells but this fraction is assumed to be low because the cells had undetectable levels of Rumi protein. (A) Control (Ras7) cells have a diagnostic band corresponding to the wild-type rumi gene (1832 bp), whereas, each of the 7 Rumi lines (R-1 through R-7) have a 344 bp band diagnostic for the rumi26 deletion. This shows that most cells in the Rumi lines are homozygous for rumi26. (B) In order to detect rare non-homozygous mutant cells, another PCR test was performed using primers that only amplify the wild-type gene. Both control Ras7 and Rumi line 3 cells had the expected 615 bp fragment. This shows that some fraction of cells in Rumi line-3 have a wild type rumi gene. (C) Western blotting was used to analyze Rumi protein in control (Ras7) and Rumi line 3 and 6 cells (R3 and R6). Only the control cells expressed detectable levels of Rumi protein. Actin was used as a control to show that similar levels of protein were loaded.