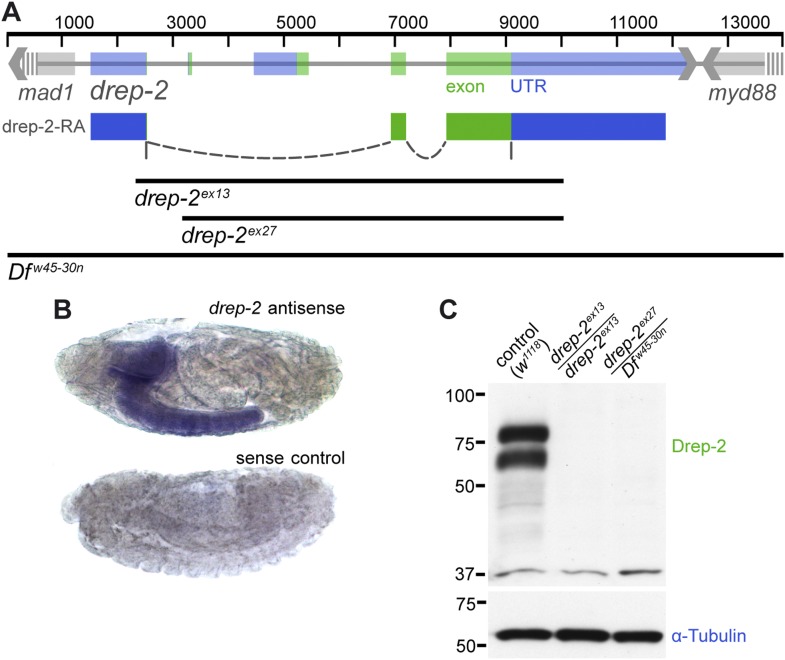
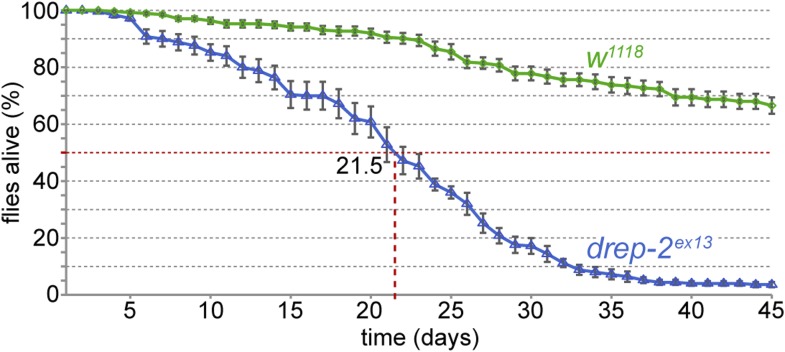
Figure 1. Expression and mutants of drep-2.
(A) Genetic scheme of the drep-2 locus on chromosome IIR. The neighboring genes mad-1 and myd88 extend beyond the sequence displayed. The cDNA labeled drep-2-RA was used for rescue experiments. Blue: untranslated regions; green: exons; black lines: deleted regions in the mutants. (B) In situ hybridization of drep-2 reveals a neuronal expression pattern (stage 17). (C) Western blot of adult fly head extracts using the anti-Drep-2C-Term antibody. Drep-2 isoforms are predicted to run at 52 and 58 kDa. The signal is absent in both the drep-2ex13 and the drep-2ex27/Dfw45-30n mutant.