Abstract
MicroRNAs are regulatory factors that play important roles in tumor development, invasion and metastasis. Previously, we showed that miR-199a is abnormally expressed in clinical melanoma specimens and expression was closely associated with clinical features of metastasis. However, the exact molecular mechanisms by which miR-199a-5p influences melanoma invasion and metastasis remains unclear. In this study, we investigated gene expression changes of metastasis-associated genes in B16F10 melanoma cells following targeted silencing or overexpression of miR-199a-5p, using mouse tumor metastasis PCR arrays. Comparison of gene expression changes in miR-199a-5p-silenced versus overexpressing cells identified a set of upregulated genes (> 2-fold) including Cd44, Cdh1, Cxcr4, Etv4, Fxyd5, Rpsa, Mmp3, Myc, Rb1, Tcf20, Hprt1, Actb1 and downregulated genes (> 2-fold) including Ctsk, Itga7 and Tnfsf10. Regulation of a subset of these genes (Myc, Tnfsf10 and Cd44) following miR-199a-5p silencing or overexpression was validated by reverse transcription-polymerase chain reaction (RT-PCR) and western blot. In conclusion, our study demonstrates that miR-199a-5p regulates melanoma metastasis-related genes, and may provide a basis for the development of novel, molecularly targeted drugs.
Keywords: Melanoma, miR-199a-5p, metastasis PCR array
Introduction
Melanoma is a malignant tumor initiated from neural crest cells and often occurs in the skin, with a clear familial aggregation [1]. Melanomas are prone to invasion and metastasis and are associated with a high mortality rate. In the early stages of disease, melanoma patients exhibit mild symptoms, with regional lymph node metastasis and even distant metastasis often visible in confirmed cases of melanoma, associated with a poor prognosis. Therefore, a deeper understanding of the specific pathways underlying melanoma cell invasion and metastasis will likely have far-reaching significance for the treatment of patients with melanoma.
Recent studies have shown that microRNAs (miRNAs) are important regulatory factors involved in the development, invasion and metastasis of melanoma [2]. MiR-18b has been shown to target MDM2-TP53 signaling [3], miR-7-5p partially inhibits IRS-2 expression and AKT signaling [4]. And miR-9 can be targeted to regulate NF-κB1 signaling, thereby inhibiting melanoma cell proliferation, invasion and metastasis [5]. Conversely, miR-182 regulates MATF-M and FOXO3; [6] and miR-21 can downregulate PTEN and PDCD4 mRNA expression and BTG2 protein expression, promoting proliferation, invasion and metastasis of melanoma [7]. However, the specific molecular mechanisms by which miRNAs may affect tumor invasion and metastasis in melanoma remain unclear.
Our previous miRNAs microarray analyses of clinical melanoma specimens revealed abnormal expression of miR-199a, and this was closely positively related to clinical features of melanoma metastasis. In the present study, a mouse tumor metastasis PCR array was used to investigate expression changes of 84 known tumor metastasis-associated genes, following targeted inhibition or overexpression of miR-199a-5p in B16F10 melanoma cells. These results were subsequently validated by RT-PCR and western blot analysis. Understanding the specific molecular mechanisms by which miR-199a-5p regulates melanoma invasion and metastasis will provide new strategies for the development of novel, molecularly targeted drugs with the ultimate goal of improving patient survival.
Materials and methods
Cell culture
The highly metastatic, B16F10 mouse melanoma cell line was purchased from Nanjing KeyGen Biotech Limited Company (China). Cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin, and incubated in a 5% CO2 humidified incubator at 37°C.
Antibodies and reagents
Myc, Tnfsf10 and Cd44 antibodies were purchased from Abgent Company (San Diego, CA, USA). The mmu-miR-199a-5p mimics and mmu-microRNA-199a-5p inhibitor plasmid was purchased from RiboBio Biological Technology Company (China). The SuperArray PCR master mix microarray kit and Mouse Metastasis gene chip (PAMM-028A) were purchased from SuperArray (USA) and SABiosciences (USA), respectively.
Grouping
Experimental treatment groups were as follows: i) Blank group: non-treated B16F10 cells; ii) miR-199a-5p silenced group: liposome-mediated transfection of B16F10 cells with mmu-microRNA-199a-5p inhibitor, iii) miR-199a-5p overexpression group: liposome-mediated transfection of B16F10 cells with mmu-miR-199a-5p mimics and plasmid.
Transfection
B16F10 cells were seeded at a density of 1×105 cells/well (final volume 2 ml) in 6-well plates. Near-confluent (80-90%) or sub-confluent (30-50%) cells were subsequently transfected with either mmu-microRNA-199a-5p inhibitor (5 μl) or mmu-miR-199a-5p mimics (5 μl), respectively.
RT-PCR
Cells were harvested 48 h post-transfection and RNA was extracted using TRIzol solution in accordance with the manufacturer’s instructions. RNA purity and concentration were measured using a NanoDrop ND-1000(USA). Complementary DNA samples were prepared using a reverse transcription kit in accordance with the manufacturer’s instructions (K1662, Fermentas, Canada). In brief, total RNA (1 μg), oligo(dT)18 (1 μl) and diethylpyrocarbonate water (up to 12 μl) were combined in a PCR tube, incubated at 65°C for 5 min and then placed on ice for 2 min. A reaction mixture containing 5× Reaction Buffer (4 μl), RNase inhibitor (20 U/μl, 1 μl), dNTP Mix (10 mM, 2 μl) and M-MuLV Reverse Transcriptase (200 U/μl, 1 μl) in a total volume of 20 μl was then added to each tube, and reactions were incubated at 42°C for 60 min and 70°C for 5 min. PCR reactions were assembled in a final volume of 20 μl including cDNA (1 μl), Forward Primer (0.5 μM), Reverse Primer (0.5 μM) and PCRMix (10 μl). Primer sequences are as follows: Myc (NM_010849.4) 353 bp: forward: 5’ AAGGGAAGACGATGACGG 3’, reverse: 5’ TGAGCGGGTAGGGAAAGA 3’; Tnfsf10 (NM_009425.2) 257 bp: forward: 5’ GGGCATTCATTTCTCAAC 3’, reverse: 5’ AGTCCGTACTCGGCATCT 3’. Cd44 (NM_001039150.1), 763 bp: forward: 5’ CCTTGGCCACCACTCCTAAT 3’, reverse: 5’ GTGGTCACTCCACTGTCCTG 3’. All primers were synthesized by Sangon Biotech Company (Shanghai, China) and primer of GADPH was bought from Fermentas (Canada). PCR was performed using the following cycling conditions: 5 min at 95°C (one cycle), then 30 s at 94°C, 30 s at 53°C, 30 s at 72°C (30 cycles) and a final extension of 7 min at 72°C. PCR reactions (5 μl) were analyzed on a 1.5% agarose gel containing ethidium bromide for 1 h, and GAPDH was used as an internal reference gene. DNA was visualized under UV light (254 nm) and images were captured.
Real-time PCR validation of miR-199a-5p expression
RNA and cDNA were prepared as described above. Following reverse transcription, reactions were prepared using SYBR Premix Ex Taq™ (DRRO41A, Takara, Japan) in accordance with the manufacturer’s instructions. PCR reactions were performed in a 20 μl reaction volume containing cDNA (2 μl), SYBR (10 μl), 10 μM forward primer (0.4 μl), 10 μM reverse primer (0.4 μl), 50X ROX (0.4 μl) and ddH2O (6.8 μl). Real-time PCR was performed using the following cycling conditions: 95°C for 10 s (one cycle), then 95°C for 5 s, 60°C for 30 s, 95°C for 15 s, 60°C for 15 s and 95°C for 15 s (40 cycles). The relative expression of miR-199a-5p (F) was calculated as follows: F = 2-ΔΔct (where Δct1 = sample group average target gene ct value-average internal reference gene value; Δct2 = Blank group average target gene ct value - average internal reference gene value; ΔΔct = Δct1-Δct2).
Real-time PCR microarray screening
Total RNA was extracted from B16F10 tumor cells following defined treatments using TRIzol solution and RNA concentration and purity were assessed by UV spectrophotometry. Metastasis-associated genes were identified using real-time mouse tumor metastasis PCR arrays (performed by Kangchen Biotech, Shanghai, China). Differentially expressed genes ≥ 2-fold (P < 0.05) were considered significant.
Western blot
B16F10 cells were collected 48 h post-transfection and total protein was extracted as previously described. The concentration of protein extracts was measured according to the Bradford method, and extracts (30 μg/lane) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were then blocked in 5% skim milk/PBS-Tween for 1.5 h and incubated with primary antibodies murine anti-Myc, Tnfsf10 and Cd44 (1:1000) and murine anti-α-tublin (1:10 000) at 4°C overnight. After washing with PBS-Tween (3×), membranes were incubated with murine, horseradish peroxidase (HRP)-conjugated secondary antibody (1:10 000) for 1 h at room temperature, washed with PBS-Tween (3×), and signal was developed using enhanced chemiluminescence (ECL) substrate.
Statistical analysis
Data were analyzed using SPSS 13.0 (SPSS, Chicago, IL, USA), and are represented as the mean ± standard deviation (SD). A paired t-test was used to examine differences between each group. A P value < 0.05 was considered statistically significant.
Results
Silencing or overexpression of miR-199a-5p in B16F10 cells
To investigate miR-199a-5p-induced gene expression changes in B16F10 cells, we performed both loss-of-function and gain-of-function analyses following transient transfection of cells with either mmu-microRNA-199a-5p inhibitor or mmu-miR-199a-5p mimics plasmid, respectively. After 24 h, analysis of miR-199a-5p expression by real-time PCR revealed downregulation of miR-199a-5p in mmu-microRNA-199a-5p inhibitor transfected cells relative to controls (0.354-fold ± 0.025). In contrast, miR-199a-5p expression was upregulated (3.815-fold ± 0.282) in mmu-hsa-miR-199a-5p mimics transfected cells compared with controls (Figure 1). These results confirm the successful targeted silencing and overexpressing of miR-199a-5p in B16F10 cells.
Figure 1.

Targeted inhibition or overexpression of miR-199a-5p in B16F10 cells. Relative miR-199a-5p expression levels in different groups.
Tumor metastatic PCR arrays of B16F10 cells following modulation of miR-199a-5p expression
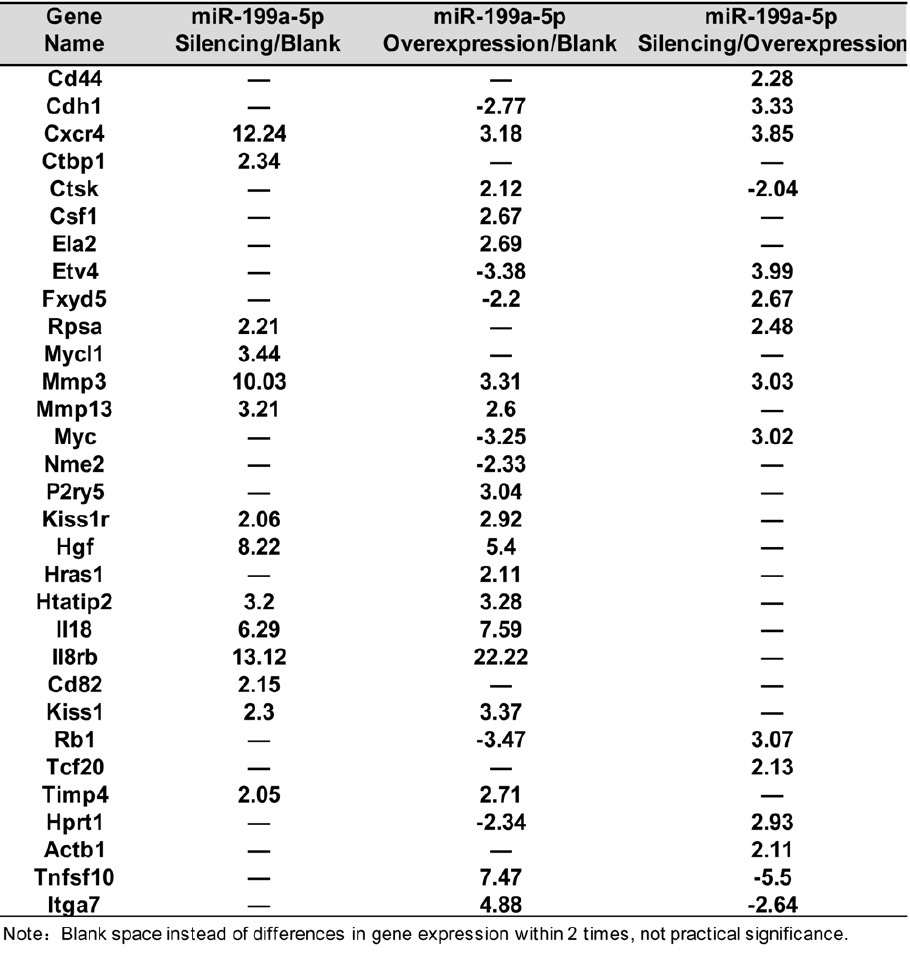
To investigate the effects of miR-199a-5p on melanoma cell invasion and metastasis, we used tumor metastasis PCR arrays to detect expression changes in 84 known tumor metastasis-associated genes in control, miR-199a-5p-silencing and miR-199a-5p-overexpressing B16F10 cells (Table 1). Real-time PCR analyses revealed that the majority of genes exhibited Ct values lower than 30, corresponding with higher gene expression (since genes with a Ct value > 35 are regarded as low/not expressed). The percentages of Ct values corresponding to < 25, 25-30, 30-35 and Absent Calls were as follows: Blank group, 67% (49), 16% (12), 11% (8), 5% (4); miR-199a-5p-silenced group, 60% (50), 14% (12), 16% (13), 10% (8) and the miR- 199a-5p-overexpressing group, 57% (46), 20% (16), 11% (14), 10% (8) (Figure 2A-C).
Table 1.
Different expressions of tumor metastatic-related genes
|
Figure 2.
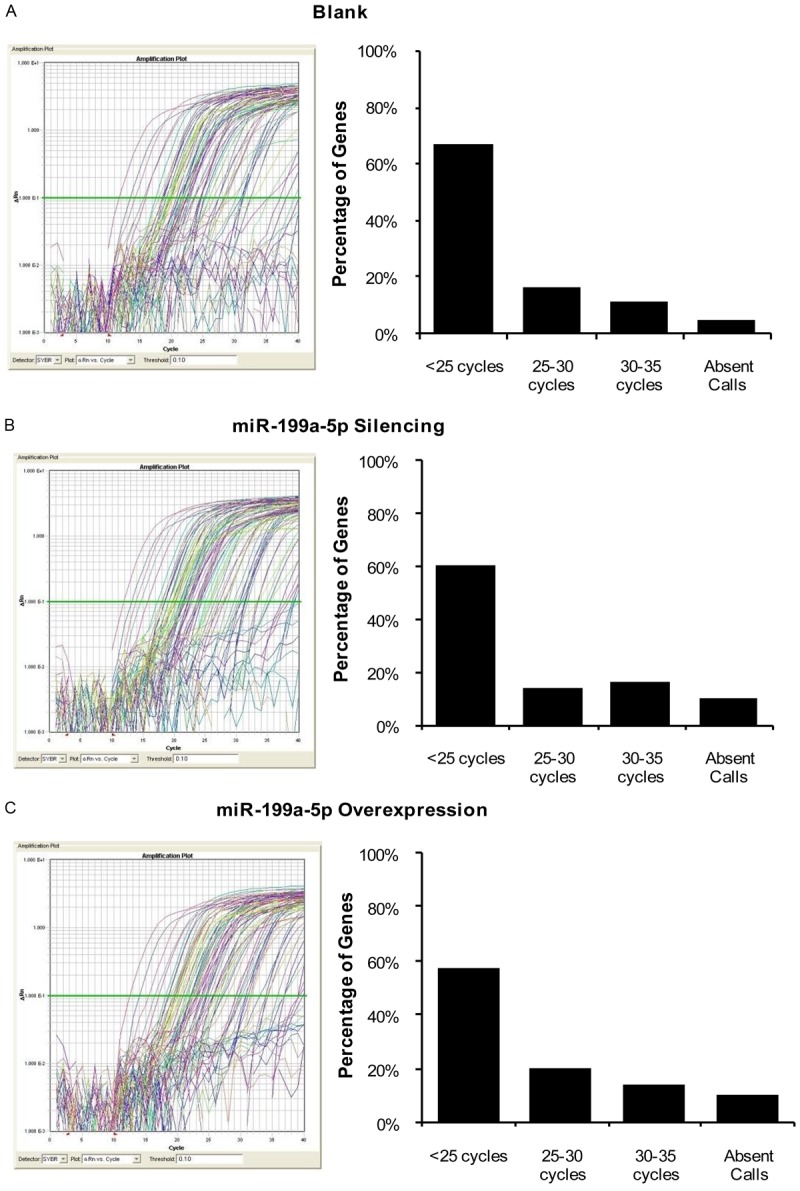
Tumor metastatic PCR arrays of WM451 cells following targeted inhibition or overexpression of miR-199a-5p. A. Amplification curves and percentage of tumor metastatic genes in the negative control (blank) cells. B. Amplification curves and percentage of tumor metastatic genes in miR-199a-5p-silenced cells. C. Amplification curves and percentage of tumor metastatic genes in miR-199a-5p-overexpressing cells.
To further investigate gene expression changes related to miR-199a-5p expression, we next investigated genes differentially expressed in miR-199a-5p-silenced cells compared with controls. We identified upregulation of 14 genes (> 2-fold) including Cxcr4, Ctbp1, Rpsa, Mycl1, Mmp3, Mmp13, Kiss1r, Hgf, Htatip2, II18, II8rb, Cd82, Kiss1 and Timp4, and there has no downregulated gene. Overexpression of miR-199a-5p led to upregulation of 17 genes (> 2-fold) including Cxcr4, Ctsk, Csf1, Ela2, Mmp3, Mmp13, P2ry5, Kiss1r, Hgf, Hras1, Htatip2, II18, II8rb, Kiss1, Timp4, Itga7, Tnfsf10, and downregulation of 7 genes, (> 2-fold) including Cdh1, Etv4, Fxyd5, Myc, Nme2, Rb1 and Hprt1. Comparison of genes differentially expressed in miR-199a-5p-silenced cells compared with overexpressing cells revealed upregulation (> 2-fold) of 12 genes including Cd44, Cdh1, Cxcr4, Etv4, Fxyd5, Rpsa, Mmp3, Myc, Rb1, Tcf20, Hprt1 and Actb1 and downregulation of 3 genes including Ctsk, Itga7 and Tnfsf10 (Table 1).
Expression of Myc, Tnfsf10 and CD44 following miR-199a-5p silencing and overexpression
To validate the accuracy of our PCR microarray results, we assessed the expression of two candidate targets, Myc and Tnfsf10, by RT-PCR and Western blot. According to the PCR microarray, comparison of gene expression between miR-199a-5p-silenced versus overexpressing cells revealed a 3.02-fold upregulation of Myc and 5.5-fold downregulation of Tnfsf10. RT-PCR and Gel-pro Analyzer image analysis revealed significant upregulation of Myc in miR-199a-5p-silenced cells compared with overexpressing cells IOD (integrated optical density) value of Myc (353 bp) to GAPDH (495 bp) of (0.72 ± 0.05) vs (0.53 ± 0.04), respectively; p < 0.05), consistent with western blot results. Conversely, Tnfsf10 expression was significantly downregulated in miR-199a-5p-silenced cells compared with overexpressing cells (IOD value of Tnfsf10 (257 bp) to GAPDH of (0.44 ± 0.03) vs (0.83 ± 0.06); p < 0.05). Furthermore, Tnfsf10 protein levels were also dramatically decreased in miR-199a-5p-silenced cells (Figure 3). Our PCR array analysis also identified upregulation of Cd44 (2.28-fold), which was previously reported to be a target of miR-199a-5p [8], in miR-199a-5p-silenced cells compared with overexpressing cells. This upregulation was subsequently validated by RT-PCR and western blot (Figure 4), further confirming the accuracy and reliability of our PCR microarray results. Thus, miR-199a-5p exerts an effect on melanoma cell invasion and metastasis by regulating the expression of multiple genes.
Figure 3.

Expression of Myc and Tnfsf10 following targeted inhibition or overexpression of miR-199a-5p. A. Expression of Myc was detected by RT-PCR and western blot. B. Expression of Tnfsf10 was detected by RT-PCR and Western blot.
Figure 4.
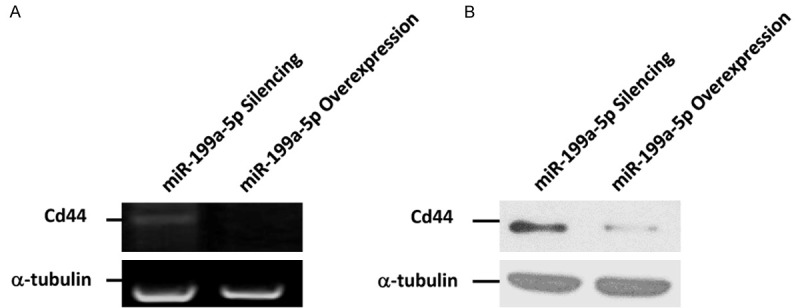
miR-199a-5p targets Cd44. A. Expression of Cd44 was detected by RT-PCR. B. Expression of Cd44 was detected by Western blot.
Discussion
miR-199a-5p with a low expression in breast cancer can be targeted by DRAM1 and Beclin1 to inhibit radiation-induced autophagy in breast cancer cells [9,10]. Low expression of miR-199a-5p has previously been observed in hepatic carcinoma [11]. Cisplatin-induced downregulation of miR-199a-5p is capable of activating autophagy of hepatoma cells, leading to drug resistance [12]. Low expression of miR-199a is also observed in ovarian cancer, targeting IKKβ and thereby affecting NF-κB activity. Furthermore, reactive oxygen species may inhibit miR-199a and miR-125b, leading to induction of ERBB2 and ERBB3 expression in ovarian cancer [13-15]. The miR-199a has also been reported as a tumor suppressor gene in renal cancer and testicular cancer in addition to other tumors [16,17]. In contrast, miR-199a may also act as an oncogene when highly expressed in gastric cancer. miR-199a is not only capable of targeting mitogen-activated protein kinase 11 [18], but also Smad4, negatively regulating tumor growth factor β signaling pathway [19]. Thus, miR-199a-5p exhibits multiple biological functions and tissue specificity.
The miR-199a is highly expressed in melanoma exhibiting high invasive ability [20]. The expression of miR-199a is increased in clinical specimens of older patients (> 60 years) with melanoma [21]. miR-199a-5p, miR-199a-3p and miR-1908 may target ApoE to drive LRP1/LRP8-dependent tumor metastasis and angiogenesis [22]. Previously, we observed that miR-199a-5p expression was remarkably correlated to the high metastatic ability of melanoma, and may represent one of several key miRNAs in melanoma metastasis [1].
To investigate the transcriptomic effect of miR-199a-5p in melanoma, we used murine tumor metastasis PCR microarray chips containing metastasis-associated genes involved in cell adhesion, extracellular matrix components, cell cycle, cell growth and proliferation, apoptosis, transcription and other biological processes. A subset of these differentially expressed genes was then validated using RT-PCR and western blot, verifying the accuracy of the PCR microarray results and validity of this screening approach. While we did not find miR-199a-5p binding target sequences in Myc and Tnfsf10 3’UTRs using TargetScan prediction software, it is possible that miR-199a-5p indirectly influences tumor invasion and metastasis-related genes, by adjusting network-related transcription factors and binding proteins. Future studies exploring the signaling pathways through which miR-199a-5p influences metastases-related genes in mouse melanoma cells are therefore necessary.
Effective control of tumor metastasis requires a deeper understanding of genes driving critical processes of invasion and metastasis in melanoma cells. To this end, miRNA interference technology has been shown to inhibit the progression of melanoma invasion [23]. Therefore, the study of miR-199a-5p and its network-based regulation of invasion and metastasis may provide a new strategy for the development of molecularly targeted therapy, with the goal of improving treatment outcome in melanoma patients.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81372140, 81301688, 81272192, 81171882, 81071645); Ph.D. Programs Foundation of Ministry of Education of China (No. 20130162110050 and 20130162120093); Program for New Century Excellent Talents in University (NCET-11-0527); Post-doctoral Foundation of Central South University (No. 131425).
Disclosure of conflict of interest
None.
References
- 1.Xu D, Tan J, Zhou M, Jiang B, Xie H, Nie X, Xia K, Zhou J. Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett. 2012;4:941–946. doi: 10.3892/ol.2012.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M, Xie H, Xiong W, Xu D, Cao K, Liu R, Zhou J, Luo C. [MicroRNA and metabolism regulation] . Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2013;38:318–322. doi: 10.3969/j.issn.1672-7347.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, Nosrati M, Sagebiel R, Miller JR 3rd, Kashani-Sabet M. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst. 2013;105:433–442. doi: 10.1093/jnci/djt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giles KM, Brown RA, Epis MR, Kalinowski FC, Leedman PJ. miRNA-7-5p inhibits melanoma cell migration and invasion. Biochem Biophys Res Commun. 2013;430:706–710. doi: 10.1016/j.bbrc.2012.11.086. [DOI] [PubMed] [Google Scholar]
- 5.Liu N, Sun Q, Chen J, Li J, Zeng Y, Zhai S, Li P, Wang B, Wang X. MicroRNA-9 suppresses uveal melanoma cell migration and invasion through the NF-kappaB1 pathway. Oncol Rep. 2012;28:961–968. doi: 10.3892/or.2012.1905. [DOI] [PubMed] [Google Scholar]
- 6.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286:39172–39178. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. Febs J. 2012;279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, Ma S, Liu X. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett. 2013;587:436–443. doi: 10.1016/j.febslet.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, Beckebaum S. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F, Xia Q. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423:826–831. doi: 10.1016/j.bbrc.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T, Mor G. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712–4723. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Xu Q, Jing Y, Agani F, Qian X, Carpenter R, Li Q, Wang XR, Peiper SS, Lu Z, Liu LZ, Jiang BH. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep. 2012;13:1116–1122. doi: 10.1038/embor.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukigi M, Bilim V, Yuuki K, Ugolkov A, Naito S, Nagaoka A, Kato T, Motoyama T, Tomita Y. Re-expression of miR-199a suppresses renal cancer cell proliferation and survival by targeting GSK-3beta. Cancer Lett. 2012;315:189–197. doi: 10.1016/j.canlet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Cheung HH, Davis AJ, Lee TL, Pang AL, Nagrani S, Rennert OM, Chan WY. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene. 2011;30:3404–3415. doi: 10.1038/onc.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song G, Zeng H, Li J, Xiao L, He Y, Tang Y, Li Y. miR-199a regulates the tumor suppressor mitogen-activated protein kinase kinase kinase 11 in gastric cancer. Biol Pharm Bull. 2010;33:1822–1827. doi: 10.1248/bpb.33.1822. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL, Zhao ZH, Zheng XF, Yang X. Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-beta signalling pathway. Nucleic Acids Res. 2012;40:9286–9297. doi: 10.1093/nar/gks667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18:184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 21.Jukic DM, Rao UN, Kelly L, Skaf JS, Drogowski LM, Kirkwood JM, Panelli MC. Microrna profiling analysis of differences between the melanoma of young adults and older adults. J Transl Med. 2010;8:27. doi: 10.1186/1479-5876-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh C, Segura MF, Gaziel-Sovran A, Menendez S, Darvishian F, Chiriboga L, Levin B, Meruelo D, Osman I, Zavadil J, Marcusson EG, Hernando E. Efficient in vivo microRNA targeting of liver metastasis. Oncogene. 2011;30:1481–1488. doi: 10.1038/onc.2010.523. [DOI] [PubMed] [Google Scholar]


