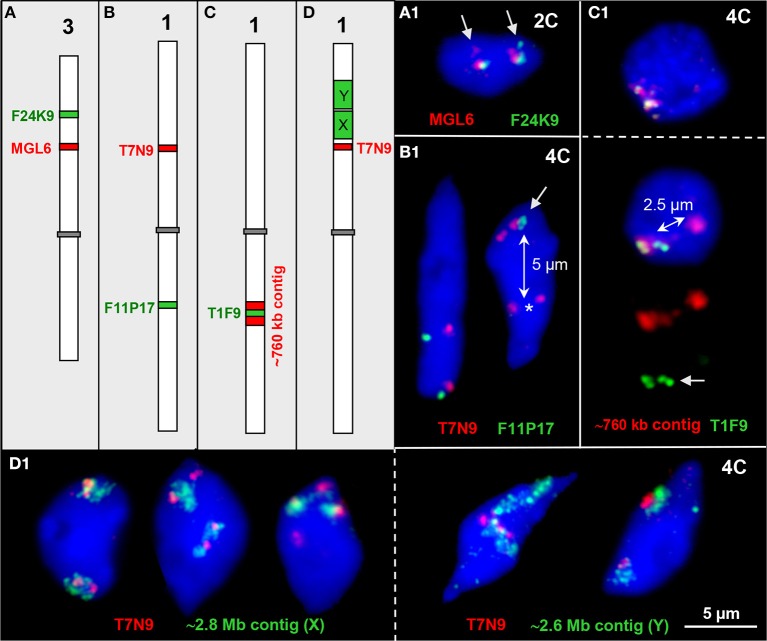
Figure 3.
Interstitial chromatin associations. (A–D) Schemes of chromosomes 1 and 3 showing interstitial positions of single BAC and BAC contig inserts in different color as used for FISH. (A1) Parts of both ~100 kb euchromatin segments are elongated in one homolog each thus showing a second signal (arrows) in an 2C nucleus. (B1) The ~100 kb segments T7N9 and F11P17 located on different arms of chromosome 1 may both be cohesive (left) or one may be separated (right, T7N9) in 4C nuclei. In the right nucleus all four F11P17 sister chromatid segments are cohesive and associated (arrow) indicated by the location apart (~5 μm, double arrow) from the lower located homolog position marked by the two T7N9 signals (asterisk). (C1) The top 4C nucleus contains the 760 kb contig of both homologs in close vicinity. The bottom nucleus displays the same contig, but the T1F9 segment of one homolog moved apart from its contig position (~2.5 μm, double arrow) toward the second homolog (arrow). (D1) Examples of chromatin configurations in 4C nuclei labeled by ~2.8 Mb (X) and ~2.6 Mb BAC (Y) contigs, respectively, in combination with the segment T7N9 from the same chromosome arm. In the first nucleus the green contigs are associated with the T7N9 segment, in the second nucleus the segments are located at the edges of the contigs, and in the third nucleus two of the non-cohesive T7N9 segments are present distantly from the contigs. The fourth and fifth nuclei demonstrate, that the T7N9 segments, although located ~3.3 Mb apart from the ~2.6 Mb contig may be localized within this domain.