FIGURE 1:
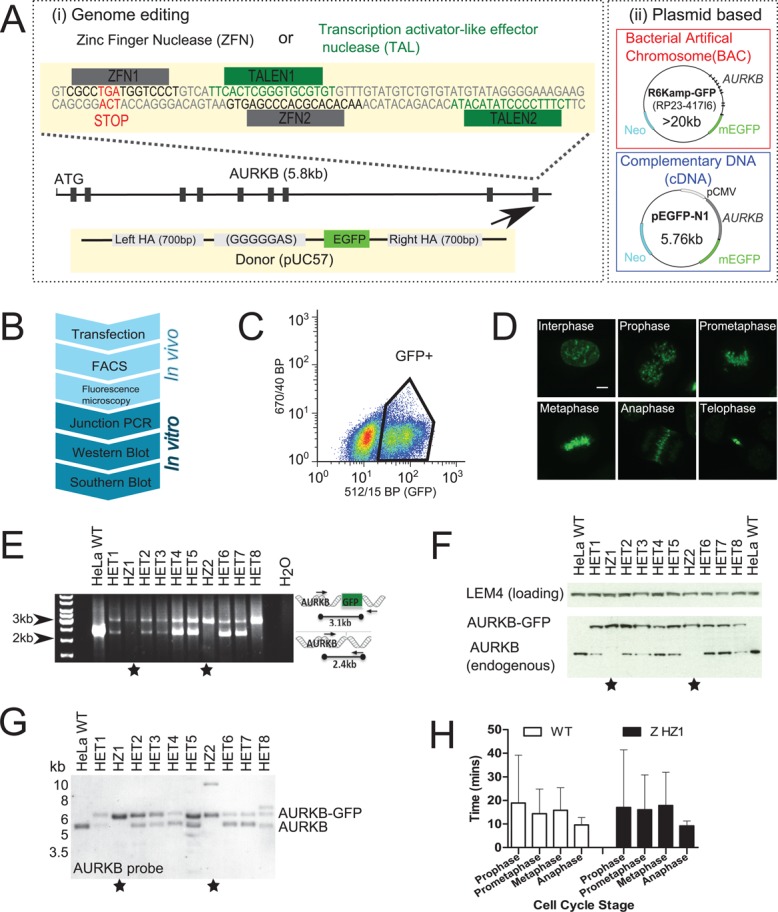
Construction and validation of genome-edited cell lines expressing AURKB-GFP. (A) Schematic of fluorescent gene–tagging systems used to create AURKB-GFP. (i) ZFNs or TALENs cause DNA double-strand breaks at the C-terminus of the AURKB locus, before repair with a donor construct containing EGFP (arrow). (ii) Plasmids containing AURKB-GFP as either the full mouse gene (BAC) or as cDNA are randomly integrated into the genome. (B) Flowchart of assays used to construct genome-edited cell lines. Junction PCR was not used to screen plasmid-based systems since the genomic locations are unknown. (C) Fluorescence-activated cell sorting of GFP-positive cells. Gates were drawn based on comparison to nonfluorescent wild-type parental cells. (D) Fluorescence confocal microscopy maximum-intensity z-projections of genome-edited AURKB-GFP cells (clone HZ2). (E) Junction PCR screening of ZFN genome-edited AURKB-GFP clonal cells. Stars denote clones with all alleles successfully targeted. (F) Western blot screening of nocodazole-arrested ZFN AURKB-GFP cells with anti-AURKB antibody. (G) Southern blot screening of ZFN AURKB-GFP cells using a probe in AURKB intron 2 after Kpnl and Nsil digestion. (H) Mitotic timing from live-cell imaging; cells were automatically classified into cell cycle stages using mCherry-H2B as previously described (Held et al., 2010). n >355 cells from two experiments.
