The expression pattern of Staufen1 during mouse skeletal muscle development is described. Sustained expression of Staufen1 in myoblasts prevents normal differentiation by causing decreases in the expression of key myogenic markers by an SMD-independent mechanism and by promoting the translational regulation of c-myc.
Abstract
Recent work has shown that Staufen1 plays key roles in skeletal muscle, yet little is known about its pattern of expression during embryonic and postnatal development. Here we first show that Staufen1 levels are abundant in mouse embryonic muscles and that its expression decreases thereafter, reaching low levels in mature muscles. A similar pattern of expression is seen as cultured myoblasts differentiate into myotubes. Muscle degeneration/regeneration experiments revealed that Staufen1 increases after cardiotoxin injection before returning to the low levels seen in mature muscles. We next prevented the decrease in Staufen1 during differentiation by generating stable C2C12 muscle cell lines overexpressing Staufen1. Cells overexpressing Staufen1 differentiated poorly, as evidenced by reductions in the differentiation and fusion indices and decreases in MyoD, myogenin, MEF2A, and MEF2C, independently of Staufen-mediated mRNA decay. However, levels of c-myc, a factor known to inhibit differentiation, were increased in C2C12 cells overexpressing Staufen1 through enhanced translation. By contrast, the knockdown of Staufen1 decreased c-myc levels in myoblasts. Collectively our results show that Staufen1 is highly expressed during early stages of differentiation/development and that it can impair differentiation by regulating c-myc, thereby highlighting the multifunctional role of Staufen1 in skeletal muscle cells.
INTRODUCTION
Skeletal muscle cell development, or myogenesis, is a tightly regulated process. Progenitor cells originating from somites are determined for the myogenic lineage and become proliferating myoblasts. On receiving proper signals, myoblasts undergo terminal differentiation by withdrawing from the cell cycle and fusing to form multinucleated myotubes. This myogenic terminal differentiation step involves the orchestrated expression of myogenic regulatory factors such as MyoD, myogenin, and myocyte enhancer factor-2 (MEF2), as well as cell cycle regulators, including p21 and c-myc (Berkes and Tapscott, 2005; Buckingham and Vincent, 2009; Bismuth and Relaix, 2010; Bentzinger et al., 2012). Although transcriptional control of myogenic events is now well recognized, posttranscriptional regulation has emerged over the years as another key level of regulation necessary for complete muscle differentiation. In particular, RNA-binding proteins such as K-homology splicing regulator protein (KSRP), Hu antigen R (HuR), CUG-binding protein (CUGBP), and muscleblind-like protein (MBNL) have been reported to play a functional role in muscle cell proliferation and differentiation by affecting mRNA stability, decay, and/or translation of key target transcripts encoding proteins essential for differentiation (Timchenko et al., 2001; Squillace et al., 2002; Figueroa et al., 2003; Briata et al., 2005; Deschenes-Furry et al., 2005; Apponi et al., 2011; Farina et al., 2012; Amirouche et al., 2013). For example, it is well known that both HuR and KSRP bind to AU-rich elements (AREs) present in the 3′ untranslated region (UTR) of specific mRNAs, yet they regulate their stability in an opposite manner. Although located predominately in the nucleus in myoblasts, HuR shuttles to the cytoplasm upon induction of differentiation, where it binds and stabilizes p21, MyoD, and myogenin mRNAs, resulting in cell cycle arrest and promotion of myogenesis (Figueroa et al., 2003; van der Giessen et al., 2003; Deschenes-Furry et al., 2005; Beauchamp et al., 2010; von Roretz et al., 2011). In parallel, KSRP, which typically destabilizes target transcripts, dissociates from the common mRNA targets p21 and myogenin, thereby also causing enhanced mRNA stability and hence acting in a coordinated manner with HuR (Briata et al., 2005).
As part of our efforts to identify RNA-binding proteins that play key roles in skeletal muscle, we became interested several years ago in Staufen (Belanger et al., 2003). Originally discovered in Drosophila, Staufen is an RNA-binding protein that associates with extensive RNA secondary structures through one or more double-stranded RNA–binding domains (Marion et al., 1999; Wickham et al., 1999). Studies in mammals revealed the existence of two genes named Staufen1 and Staufen2. The exact roles of Staufen1 have yet to be defined in mammals; however, it is known to be a component of RNA granules. In neurons, Staufen1-containing granules are transported in dendrites in a microtubule-dependent manner, suggesting that Staufen1 is also involved in mRNA transport (Kiebler et al., 1999; Kohrmann et al., 1999; Krichevsky and Kosik, 2001; Mallardo et al., 2003; Brendel et al., 2004; Kanai et al., 2004). This is supported by the observation that cultures of hippocampal neurons isolated from mice expressing a mutant form of Staufen1, which lacks the third RNA-binding domain, display impairment in the dendritic delivery of mRNA-containing ribonucleoprotein particles, as well as a clear reduction in spine morphogenesis (Vessey et al., 2008). Staufen1 assumes other functions in mammalian cells in addition to its conserved role in mRNA transport. For example, Staufen1 regulates the translational efficiency of a subpopulation of transcripts when bound to their 5′UTR (Dugre-Brisson et al., 2005). Furthermore, when Staufen1 binds downstream of a natural termination codon, it appears to elicit RNA degradation by a mechanism referred to as Staufen1-mediated mRNA decay (SMD; Kim et al., 2005). Finally, we recently discovered a novel function of Staufen1 in alternative pre-mRNA splicing (Ravel-Chapuis et al., 2012). Subsequent work has shown that Staufen1 interacts with splicing factors in 293T cells (Milev et al., 2012) and that it binds preferentially to transcripts that are alternatively spliced in Drosophila cells (Laver et al., 2013), thereby confirming our initial observation for a role of Staufen1 in pre-mRNA splicing. Altogether it is clear, therefore, that Staufen1 is a multifunctional protein involved in the regulation of distinct cellular events.
In a previous study, we characterized the skeletal muscle expression of Staufen1, as well as its preferential accumulation within the postsynaptic sarcoplasm of neuromuscular junctions (Belanger et al., 2003; see also Gardiol and St Johnston, 2014). In fact, we were the first to report that Staufen1 levels are regulated during myogenic differentiation of cultured cells and that its level of expression varies according to the state of innervation and the phenotype of muscle fibers. These initial observations suggested that Staufen1 is a key component of muscle fiber plasticity and maturation. Others later confirmed the involvement of Staufen1 in myogenic differentiation, as the depletion of Staufen1 from C2C12 myoblasts achieved by small interfering RNA promotes the spontaneous formation of myotubes in the absence of myogenesis induction (Yamaguchi et al., 2008). In this context, it was also proposed that Staufen1 regulates myogenic differentiation by participating in the balance between two competing mRNA decay pathways: SMD (see earlier discussion) and nonsense-mediated mRNA decay (NMD). More specifically, it has been proposed that upon induction of differentiation, the efficiency of SMD is increased, whereas that of NMD is reduced. As a consequence, SMD targets such as Paired box 3 (Pax3) mRNAs are destabilized, whereas classical NMD targets, that is, myogenin transcripts, are stabilized (Gong et al., 2009). More recently, it was also reported that Staufen1-binding sites can be formed by intermolecular duplexing between short-interspersed elements (SINEs) found in the 3′UTR of target transcripts and those found in some long, noncoding RNAs, which, functionally, can expand the list of putative SMD targets (Wang et al., 2013).
Despite these advances, it remains unknown whether Staufen1 is also regulated during mouse skeletal muscle development in vivo. In addition, the contribution of Staufen1 to the regulation of major myogenic regulatory factors and transcription factors such as MyoD or MEF2 remains unclear. Here we report the pattern of expression of Staufen1 during embryonic and postnatal skeletal muscle development. In addition, we complement these findings by also examining the expression profile of Staufen1 during cardiotoxin-induced muscle degeneration/regeneration, a well-described in vivo model of myogenic differentiation. Finally, to better define mechanistically the role of Staufen1 in myogenic differentiation, we generated stable muscle cell lines overexpressing Staufen1 and assessed the functional consequences of this increased expression on several markers of the myogenic program. Collectively our results show that Staufen1 is highly expressed during early stages of muscle differentiation/development and progressively decreases as differentiation proceeds, reaching low levels in mature muscle. Moreover, we show that sustained expression of Staufen1 impairs differentiation by markedly reducing expression of key myogenic regulatory factors in a SMD-independent manner while promoting translation of c-myc, which causes undifferentiated myoblasts to remain in a proliferative state.
RESULTS
Staufen1 expression decreases during mouse skeletal muscle development
To better understand the role of the RNA-binding protein Staufen1 in skeletal muscle in vivo, we sought to investigate its temporal pattern of expression during mouse skeletal muscle development. We performed Western blot analyses using protein extracts collected from wild-type mice at different stages of embryonic and postnatal development ranging from embryonic day 14.5 (E14.5) to adult (14 wk). Given the small size of limbs of mouse embryos, the whole muscle mass of the leg was used for E14.5, E18.5, and postnatal day 1 (PN1). Tibialis anterior (TA) muscles were used for adult mice.
As a control, we first examined expression of CUGBP1, a developmentally regulated RNA-binding protein in skeletal muscle (Ladd et al., 2001, 2005; Lin et al., 2006). We show that its expression is significantly decreased during skeletal muscle development (p < 0.001), recapitulating expression profiles previously observed (Figure 1, A and B). Then we analyzed Staufen1 levels and showed that Staufen1 is highly abundant in embryonic muscle limbs at E14.5 (Figure 1, A and C). However, expression of Staufen1 decreases gradually (p < 0.001) throughout skeletal muscle mass development, resulting in a low level of expression in mature adult muscle. Because the whole muscle mass was used in these experiments, we cannot exclude that Staufen1 is decreased in several cell types contained within developing muscle tissues. By contrast, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were used as loading controls and show increased expression during muscle mass development (Figure 1A). Together these results suggest that expression of the RNA-binding protein Staufen1 is developmentally regulated during mouse skeletal muscle development.
FIGURE 1:
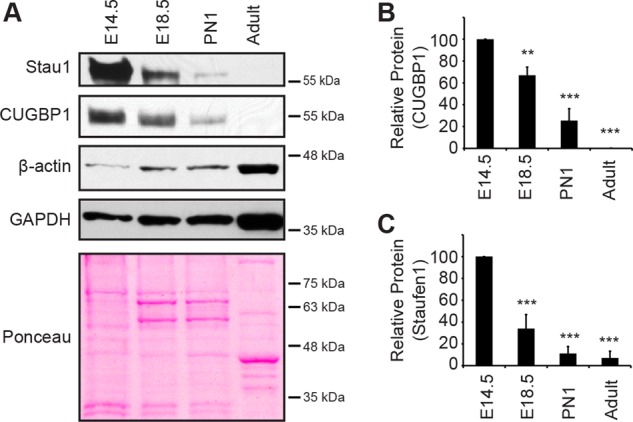
Staufen1 decreases in developing wild-type muscle. (A) Representative Western blots showing Staufen1, CUGBP1, β-actin, and GAPDH protein levels during skeletal muscle development. Samples were from embryos (E14.5 and E18.5), new-born mice (PN1), and adult mice (14 wk). Ponceau staining was used to show equal loading. (B, C) Relative quantification of Staufen1 and CUGBP1 protein levels, respectively (n = 3). Asterisks indicate significance (**p ≤ 0.01, ***p ≤ 0.001).
Expression of staufen1 is modulated during muscle regeneration
To further show that expression of Staufen1 is regulated during muscle development in vivo, we also performed muscle regeneration experiments. Briefly, we injected cardiotoxin (CTX) in wild-type adult mouse TA muscles to induce muscle degeneration. After the initial degeneration period, muscle stem cells become activated, fuse, and differentiate to repair damaged fibers and create new ones, thereby partially recapitulating characteristics of myogenesis that occur during embryonic development (Condrea, 1974; Bentzinger et al., 2012; Yin et al., 2013).
Western blots were performed using TA protein extracts obtained 2, 4, 7, and 14 d after cardiotoxin injection. Myogenin protein expression was initially analyzed. Note that in contrast to myotubes, adult muscles express low levels of myogenic regulatory factors, including myogenin, because their expression is repressed by electrical activity generated by motor neurons (Eftimie et al., 1991). Myogenin showed an increase (p < 0.001) in expression immediately after injury, thereby confirming the induction of muscle regeneration. This was followed by a steady decrease (p < 0.001 and p < 0.05) in myogenin expression levels as the regeneration process advanced to completion 14 days after cardiotoxin injection (Figure 2, A and B). As a control, GAPDH expression was measured and showed a slight decrease in protein levels at days 2 and 4 postinjury, as previously described (Orengo et al., 2011). In addition, Ponceau staining was used to confirm relatively even loading. Next we measured Staufen1 protein expression and observed that Staufen1 gradually increased (p < 0.001) from 2 to 7 d after cardiotoxin treatment (Figure 2, A and C). Staufen1 expression then returned to control levels 14 d postinjury when muscle fibers are fully regenerated. Variations of Staufen1 levels within different time points reflect interindividual variability of protein expression, which is commonly observed when using animal tissues. This induction of Staufen1 after cardiotoxin injection follows a pattern similar to the one observed with CUGBP1, which is also involved in the regulation of myogenic differentiation (Orengo et al., 2011).
FIGURE 2:
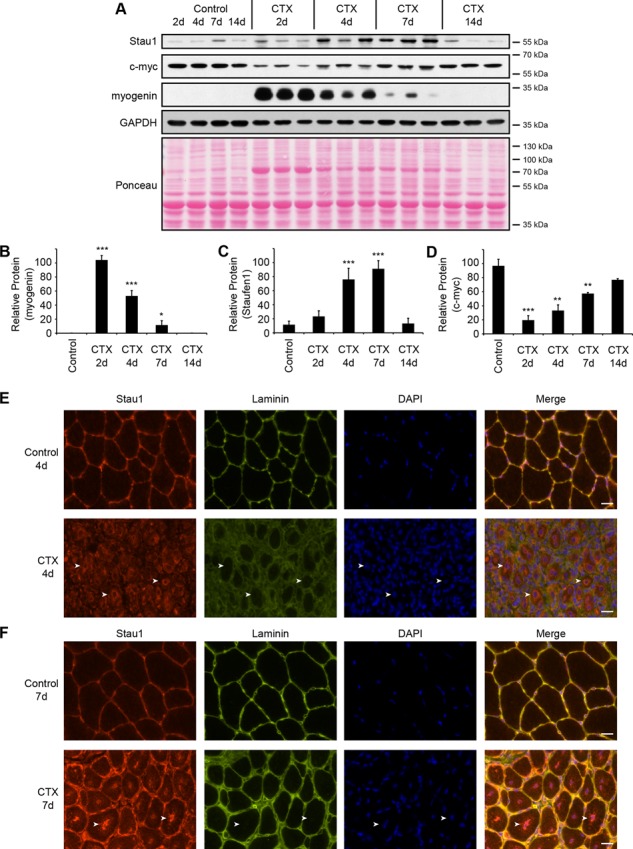
Staufen1 expression is modulated during skeletal muscle degeneration/regeneration. (A) Western blots showing Staufen1, c-myc, and myogenin protein levels in regenerating TA muscles. TA muscles were injected with CTX to induce muscle degeneration and harvested at 2, 4, 7, and 14 d postinjection. Saline-injected muscles were used as controls. GAPDH and Ponceau staining were used to show equal loading. (B–D) Relative quantification of myogenin, Staufen1, and c-myc expression levels, respectively (n = 3). (E, F) Immunofluorescence on cryostat cross sections of control and CTX-injected TA muscles. Sections were stained with a Staufen1 (red) and laminin (green) antibodies and nuclei with DAPI (blue). Arrows point to marked staining of Staufen1. Same exposure parameters were used to allow the comparison of signal intensity. Using these parameters, a no-primary-antibody control shows no signal. Scale bars, 20 μm. Asterisks indicate significance (*p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001).
To determine the cell type in which Staufen1 is increased during regeneration, immunofluorescence experiments were performed using Staufen1 antibodies on cryostat cross sections of TA muscles obtained 4 and 7 d after cardiotoxin injections and control, saline-injected muscles. Laminin was used to delineate individual muscle fibers. In normal mature fibers, Staufen1 is expressed at low levels, but faint cytoplasmic staining at the subsarcolemma can be distinguished (Figure 2, E and F). In sharp contrast, Staufen1 levels are markedly increased in the cytoplasm of muscle fibers 4 d after cardiotoxin injection, mostly in the perinuclear region of centrally located nuclei (Figure 2E). Such relocalization of Staufen1 during muscle regeneration likely reflects a need for an enhanced nuclear function of Staufen1 at this regeneration step. At 7 d postinjury, Staufen1 partially returns to the periphery of muscle fibers but remains strongly associated with nuclei and perinuclear regions (Figure 2F). Therefore our results clearly show an increase of Staufen1 levels within regenerating skeletal muscles fibers, thereby further indicating that Staufen1 is regulated during mouse skeletal myogenesis in vivo.
Levels of Staufen1 also decrease during myogenic differentiation of cultured cells
Primary human skeletal muscle cells (SkMCs) and myoblasts (HSMM) isolated from normal donors were also used to assess expression of Staufen1 during myogenic differentiation. The switch from high to low serum culture conditions induces myoblasts to exit the cell cycle and initiates differentiation by promoting the fusion of primary myoblasts into elongated, multinucleated myotubes. We thus measured the level of Staufen1 protein expression by Western blot during human primary cell differentiation and observed a progressive decrease in Staufen1 levels as cells differentiate (Figure 3, A and B). In fact, Staufen1 levels steadily decreased within 1–4 d after the switch to low serum, a time corresponding to the engagement of cells into the differentiation process, as confirmed by the induction of myogenin (Figure 3A).
FIGURE 3:
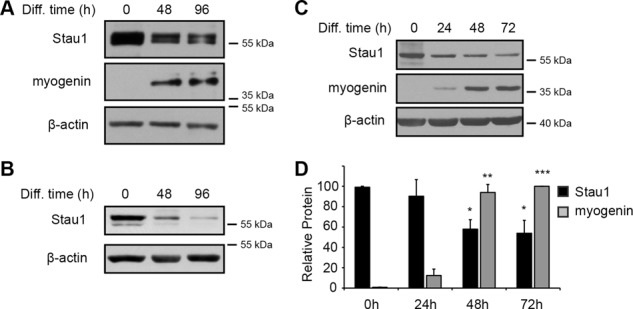
Staufen1 decreases during myogenic differentiation. (A, B) Western blots showing Staufen1 and myogenin protein levels in differentiating SkMCs and HSMM, respectively. β-Actin was used as a loading control. (C) Representative Western blots showing Staufen1 and myogenin protein levels in differentiating C2C12 cells. β-Actin was used to show equal loading. (D) Relative quantification of Staufen1 and myogenin protein levels from differentiating C2C12 cells, respectively (n = 3). Asterisks indicate significance (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
Mouse C2C12 myoblasts consist of a myogenic cell line derived from satellite cells, which have been extensively used over the years as a model system to study myogenic differentiation ex vivo (Yaffe and Saxel, 1977). We were the first to report that Staufen1 is expressed and regulated in skeletal muscles (Belanger et al., 2003). However, after this initial discovery, discrepancies in the levels of Staufen1 protein and the role of Staufen1 during the myogenic differentiation were reported (Kim et al., 2007; Yamaguchi et al., 2008, 2012; Gong et al., 2009). Conflicting data and conclusions over time, even within articles from the same laboratories, led us to reanalyze the level of Staufen1 in these cells. We therefore also measured the levels of Staufen1 and myogenin proteins during C2C12 differentiation (Figure 3, C and D) and observed a steady decrease of Staufen1 (p < 0.05), whereas myogenin is induced (p < 0.001). This decreasing pattern of Staufen1 expression in pure myogenic cell populations derived from both human and mouse as cells mature parallels the patterns observed during embryonic muscle development and muscle regeneration. Together the latter findings indicate that the patterns of Staufen1 expression observed in vivo reflect, at least partially, the modulation of Staufen1 levels within muscle cells.
Increased expression of Staufen1 reduces the efficiency of myogenic differentiation
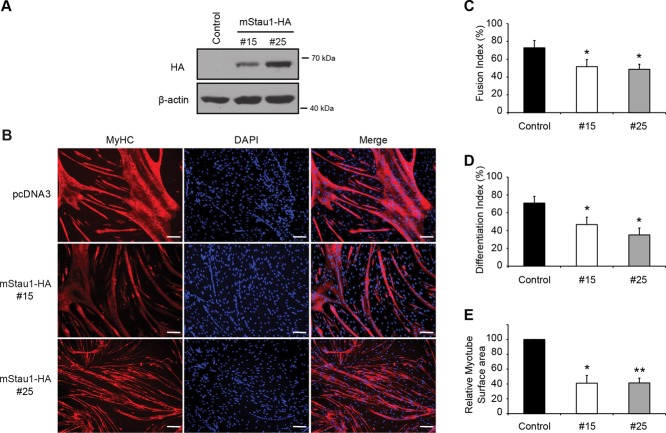
To determine specifically the effect of Staufen1 in skeletal muscle cell development and investigate its involvement in myogenic control, we sought to prevent the decrease of Staufen1 by establishing stable C2C12 myoblast clones that overexpress a hemagglutinin (HA)-tagged Staufen1 construct. Stable C2C12 myoblasts were obtained by selecting for neomycin resistance with G418. Neomycin-resistant clones were then screened for Staufen1-HA expression by Western blotting using an anti-HA antibody (Figure 4A). Two clones, #15 and #25, were selected, along with a control cell line established with a pcDNA3 control vector. Comparative analysis between control and Staufen1-HA #25 was performed by Western blot using anti-Staufen1 antibodies and showed that Staufen1-HA is overexpressed by at least ∼40% in clone #25 (unpublished data).
FIGURE 4:
Staufen1 overexpression inhibits myogenic differentiation. (A) Representative Western blots showing expression of Staufen1-HA in stable C2C12 cell lines. Two clones displaying transgene expression were chosen (#15 and #25), along with a control stable cell line. (B) Immunofluorescence of stable cell lines after 96 h of differentiation. Cells were stained with a pan-MyHC antibody (red) and nuclei with DAPI (blue). Scale bars, 100 um. (C) Fusion index (n = 5), (D) differentiation index (n = 5), and (E) myotube surface area (n = 3) of stable cell lines after 96 h of differentiation. Asterisks indicate significance (*p ≤ 0.05, **p ≤ 0.01).
To test the effect of Staufen1-HA overexpression, we initially performed morphological analyses to assess the kinetics and efficiency of differentiation of the stable cell lines. Fusion of myoblasts into myotubes in both clones #15 and #25 was substantially reduced at 96 h of differentiation, as shown by immunofluorescence using a pan–myosin heavy chain (MyHC) antibody (Figure 4B). Decreased myoblast fusion likely results from a defect in the ability of cells to fuse, but it can also be caused by a decrease in the differentiation potential of myoblasts. To further quantify this defect in differentiation, we determined the fusion index (percentage of nuclei within myotubes having ≥3 nuclei) and differentiation index (percentage of nuclei within myotubes plus MyHC-positive mononucleated cells). Both fusion and differentiation indices were significantly (p < 0.05) decreased in clones #15 and #25 at 96 h of differentiation (Figure 4, C and D). In addition, the decrease in fusion and differentiation indices was accompanied by a reduction (p < 0.05) in the overall myotube surface area (Figure 4E).
To complement these morphological analyses and further characterize the differentiation defects, we analyzed expression of two key muscle genes expressed during the early and later stages of muscle differentiation. Protein and mRNA expression were determined via Western blot and real-time quantitative reverse transcription-PCR (qRT-PCR), respectively. Our results show that overexpression of Staufen1-HA induced a dramatic reduction (p < 0.001) in the expression of myogenin and MyHC in clones #15 and #25 as assessed by Western blotting (Figure 5, A–C). The decrease in myogenin expression was also confirmed (p < 0.05) at the transcript level by qRT-PCR (Figure 5D).
FIGURE 5:
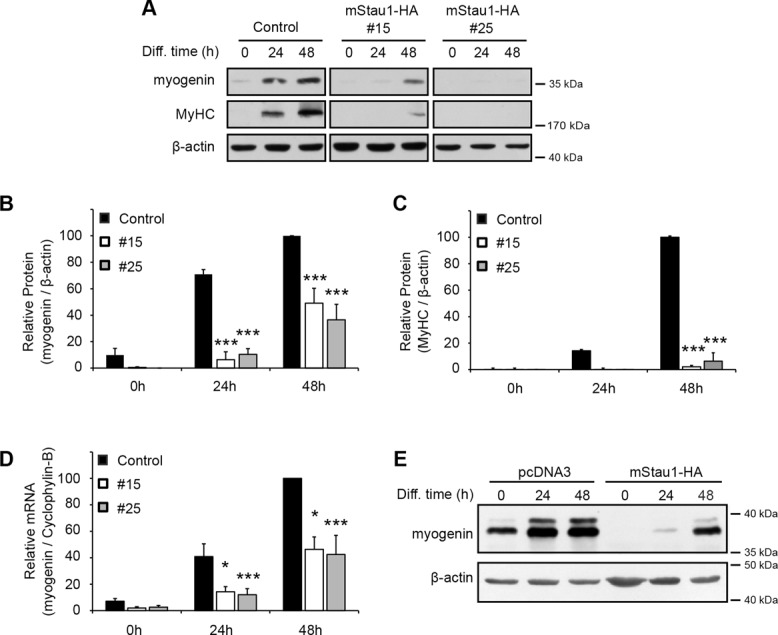
Staufen1 decreases the expression of the myogenic markers myogenin and MyHC. (A) Representative Western blots showing myogenin and MyHC expression during differentiation of Staufen1-HA–stable C2C12 cells. β-Actin was used to show equal loading. (B, C) Relative quantification of myogenin and MyHC protein levels, respectively (n = 3). (D) Relative quantification of myogenin mRNA levels as determined by qRT-PCR (n = 4). Levels were normalized to cyclophylin-B. (E) C2C12 cells were transiently transfected with mouse Staufen1-HA or a control empty vector (pcDNA3). At 24 h after transfection, cells were switched to differentiation medium and differentiated for the indicated time. Western blots showing myogenin expression in transfected cells. β-Actin was used as a loading control. Asterisks indicate significance (*p ≤ 0.05, ***p ≤ 0.001).
C2C12 myoblasts can spontaneously lose the ability to differentiate and fuse into myotubes due to selection through successive passages. In addition, selection of individual stable clones can result in the selection of myoblasts with altered intrinsic differentiation potential. Therefore, to ensure that phenotypic and molecular changes observed in clones #15 and #25 are due to the expression of exogenous Staufen1-HA, transient transfections were also performed (Figure 5E). The results obtained from transient transfections also showed pronounced reduction in myogenin expression in Staufen1-HA–overexpressing cells, which significantly supports and reproduces findings obtained with stable cell lines, thereby ruling out a clonal effect.
The decrease in differentiation potential is MyoD independent
We then sought to investigate whether Staufen1 regulates expression of the myogenic regulatory factor MyoD, a crucial regulator of myogenesis (Tapscott et al., 1988; Berkes and Tapscott, 2005; Buckingham and Vincent, 2009; Bismuth and Relaix, 2010; Bentzinger et al., 2012). We found that during proliferation and differentiation, expression of MyoD was significantly reduced (p < 0.05) in both Staufen1-HA–overexpressing clones compared with control cells (Figure 6, A–C). To assess whether the addition of exogenous MyoD could rescue the differentiation defect, we performed MyoD add-back experiments. Briefly, stable cell lines were transfected with MyoD-Flag-myc or control vectors. After transfection, cells were allowed to differentiate for 3 d. First, the increase in MyoD expression was confirmed by Western blot using MyoD antibodies. In these blots, the lower band corresponds to endogenous MyoD and the upper band to ectopic MyoD-Flag-myc (Figure 6D). Note that we were able to restore normal MyoD levels in Staufen1-HA stable cell lines (compare CTL+pcDNA3 and #25+MyoD). Then we assessed the efficiency of differentiation by measuring expression of myogenin and by immunofluorescence using a pan-MyHC antibody. As illustrated in Figure 6, E–G, no rescue of differentiation was observed in the Staufen1-HA stable cell line upon addition of MyoD-Flag-myc (compare both Staufen1-HA cells transfected with MyoD-Flag-myc to control cells transfected with MyoD-Flag-myc and to Staufen1-HA cells without MyoD-Flag-myc). Taken together, these findings show that the inhibition of differentiation induced by Staufen1 is MyoD independent.
FIGURE 6:
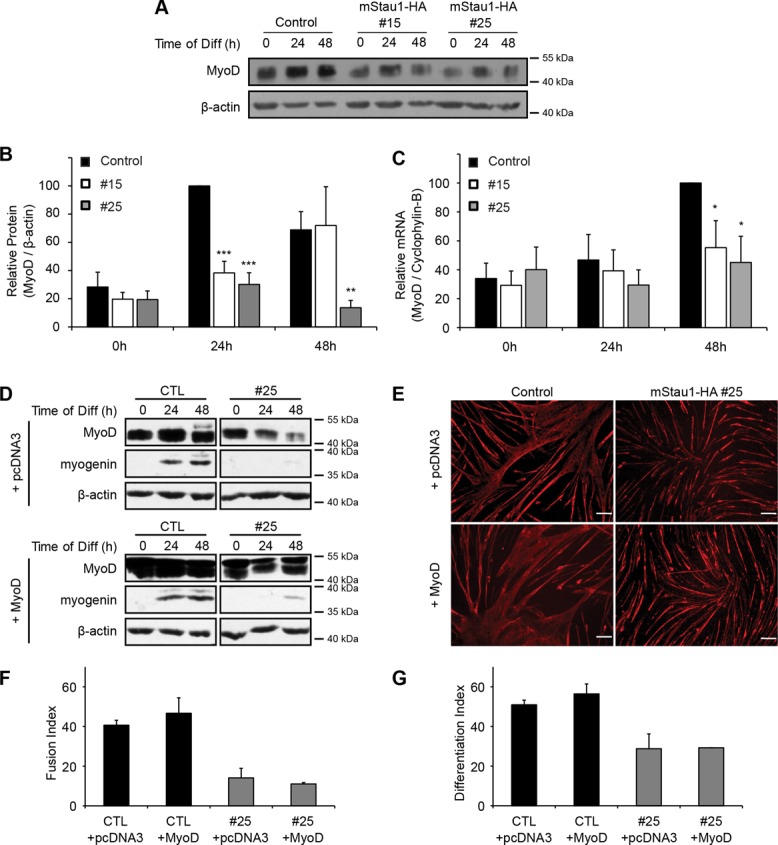
MyoD does not rescue the differentiation defect. (A) Representative Western blots showing MyoD protein level during myogenic differentiation of Staufen1-HA–stable cell lines. β-Actin was used as a loading control. (B) Relative quantification of MyoD protein levels normalized to β-actin (n = 4). (C) Relative quantification of MyoD mRNA levels as determined by qRT-PCR. Levels were normalized to cyclophylin-B (n = 4). (D) Stable cell lines were transfected with control or MyoD expression vectors. At 24 h after transfection, cells were allowed to differentiate for the indicated time period. Western blots showing MyoD (lower band, endogenous MyoD; upper band, ectopic MyoD-Flag-myc protein) and myogenin expression levels. β-Actin was used to show equal loading. (E) Immunofluorescence using a pan-MyHC antibody. (F, G) Fusion and differentiation indices of C2C12 cells differentiated for 72 h. Scale bars, 100 μm. Asterisks indicate significance (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
Overexpression of Staufen1 decreases levels of MEF2 mRNAs in a SMD-independent manner
It is well known that the transcription factor MEF2 synergizes with MyoD to control the myogenic program (Lilly et al., 1994; Molkentin et al., 1995; Berkes and Tapscott, 2005; Buckingham and Vincent, 2009; Bismuth and Relaix, 2010; Bentzinger et al., 2012). We therefore examined the mRNA levels of MEF2A and MEF2C by qRT-PCR in the Staufen1-HA–overexpressing clones. MEF2A and MEF2C mRNA levels were both decreased (p < 0.05) in Staufen1-HA–overexpressing clones compared with those seen in control cells (Figure 7, A and B). In these experiments, we observed a slightly greater variability in MEF2C mRNA levels within conditions. This likely explains why MEF2C mRNA levels are significantly decreased only at 48 h in clone #15. As mentioned in the Introduction, Staufen1 is believed to be involved in the control of mRNA stability by a mechanism referred to as Staufen1-mediated mRNA decay, which involves direct binding of Staufen1 to secondary structures present in the 3′UTR of target mRNAs, and subsequent mRNA decay (Kim et al., 2005). This degradation mechanism has been shown to target a specific subset of mRNAs, including Pax3, whereas other transcripts, such as myogenin, are unaffected (Gong et al., 2009).
FIGURE 7:
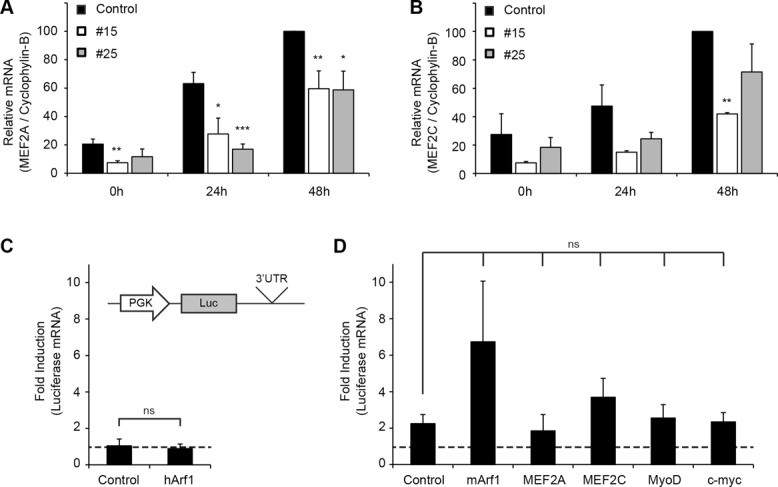
MEF2A, MEF2C, MyoD, and c-myc are not SMD targets. (A, B) Relative quantification of MEF2A and MEF2C mRNA levels in stable C2C12 cells as determined by qRT-PCR. Levels were normalized to cyclophylin-B (n = 4 and 3, respectively). (C) HeLa cells were cotransfected with luciferase vectors containing control or human Arf1 3′UTR together with human Staufen1. After transfection, relative levels of luciferase mRNAs were determined by qRT-PCR and normalized to 18S. Data were also normalized to the level of luciferase mRNAs in absence of Staufen1 overexpression. (D) HeLa cells were cotransfected with luciferase vectors containing control or mouse Arf1, MEF2A, MEF2C, MyoD, or c-myc 3′UTRs together with mouse Staufen1. Data were analyzed as in C (n = 3). Asterisks indicate significance (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001); ns, not significant.
We thus wondered whether Staufen1 induces decay of other mRNAs involved in myogenic differentiation in Staufen1-HA–overexpressing stable cell lines. In this context, MEF2A mRNA appeared as a good candidate, as it was found to be present in Staufen1-containing ribonucleoproteins, as revealed by microarray analysis (Furic et al., 2008), suggesting that Staufen1 may directly bind to and regulate MEF2A mRNA. Thus we examined whether MEF2A and MEF2C are targets of SMD. To test this, we used a similar assay to the one previously described in the initial description of SMD (Kim et al., 2005). Briefly, luciferase constructs containing the 3′UTR of potential targets were generated (Figure 7, C and D) and transfected in HeLa cells. If these transcripts are SMD targets, cotransfection of these 3′UTR-luciferase constructs together with Staufen1-HA should result in a decrease in reporter mRNA expression. Our results demonstrate that Staufen1-HA overexpression did not induce decay of MEF2A, MEF2C, and MyoD reporter constructs, thereby ruling out a contribution of SMD to the lower levels of these myogenic regulatory factors observed in our stable cell lines (Figure 7D). It is important to note that similar results were obtained in C2C12 myoblasts (unpublished data). Given these negative results, we decided to also examine as a positive control whether indeed Staufen1 can decrease expression of luciferase-3′UTR constructs containing the Arf1 3′UTR as originally done in the work leading to the concept of SMD (Kim et al., 2005, 2007). In our hands, Arf1 from both humans (Figure 7C) and mice (Figure 7D) did not appear subject to SMD (see Discussion for more details).
Overexpression of Staufen1 promotes c-myc translation and myoblast proliferation
In parallel to the characterization of myogenic regulatory factors involved in muscle differentiation, we also performed a targeted approach to identify mRNAs regulated by Staufen1 and responsible for the impaired myogenesis. Although we examined the potential role of several candidates, including p21, RalGDS, cdk5, p35, junD, and c-jun, we were particularly interested in the transcription factor c-myc because it plays a role in the balance between proliferation/differentiation and is a known potent myogenic inhibitor (Miner and Wold, 1991). Thus we analyzed the levels of c-myc protein and mRNA by Western blot and qRT-PCR, respectively, in control versus Staufen1-HA–stable cell lines. In these experiments, we observed a significant increase (p < 0.05) in c-myc protein levels in Staufen1-overexpressing cells without a parallel increase in mRNA expression (Figure 8, A–C). In addition, we next determined whether knocking down Staufen1 expression had a reverse effect on c-myc protein levels in muscle cells. Thus C2C12 cells were transiently transfected with a short hairpin RNA (shRNA) targeting Staufen1 or a control. Western blot analyses showed a slight knockdown of Staufen1 in these experiments, together with a modest decrease in c-myc protein expression (Figure 8D). To circumvent the limitations linked to the relatively low transfection efficiency of C2C12 cells and improve the extent of the Staufen1 knockdown, we also transduced human primary HSMM cells with lentiviruses expressing one of two different shRNAs targeting Staufen1 or a control lentivirus. Western blots showed that both shRNAs induced a marked reduction in Staufen1 expression. As expected on the basis of our overexpression studies, down-regulation of Staufen1 was accompanied by a decrease in c-myc protein levels (Figure 8E). Taken together, data obtained with these varied experimental systems show that Staufen1 clearly regulates c-myc protein levels in skeletal muscle cells. Finally, to complement this work, we also assessed the levels of c-myc during muscle regeneration. After an initial decrease in c-myc levels at 2 d after cardiotoxin injection, the level of c-myc increased and essentially mirrored that of Staufen1 during early to mid phases of muscle regeneration, as expected based on our work with cells in culture (Figure 2, A and D).
FIGURE 8:
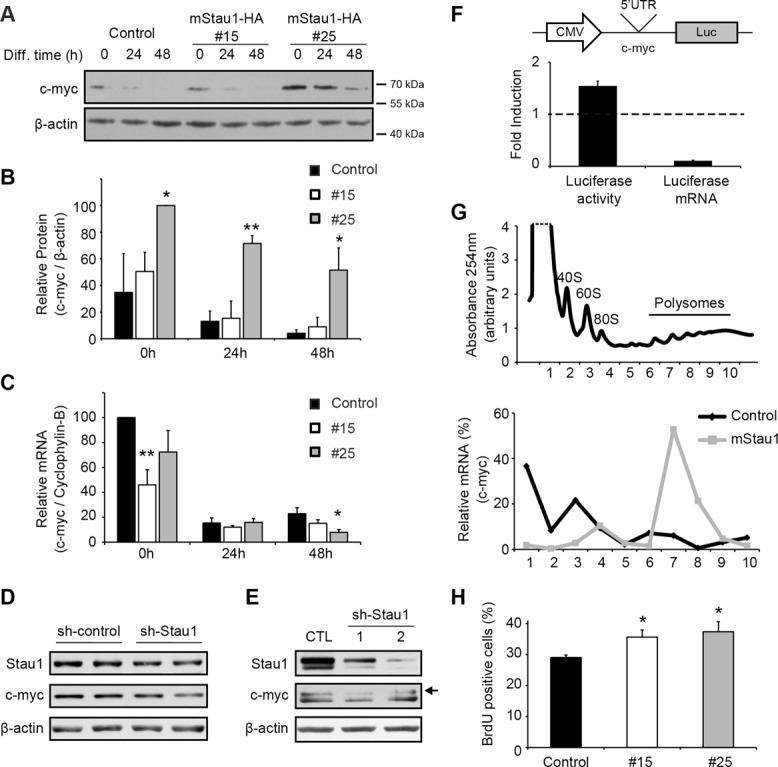
Staufen1 overexpression increases c-myc protein expression levels and cell proliferation. (A) Representative Western blot showing c-myc protein levels during myogenic differentiation of stable C2C12 cells. β-Actin was used to show equal loading. (B) Relative quantification of c-myc protein levels normalized to β-actin (n = 3). (C) Relative quantification of c-myc mRNA level in stable cell lines as determined by qRT-PCR. Levels were normalized to cyclophylin-B (n = 3). (D) C2C12 cells were transfected with sh-Staufen1 or control vectors. Western blots showing decreased Staufen1 and c-myc protein levels. β-Actin was used to show equal loading. (E) HSSM primary cells were transduced with sh-Staufen1 or control lentiviruses. Western blots were performed as in D. The arrow shows the specific c-myc band, as determined in separate experiments by knocking down c-myc expression (unpublished data). (F) 293T cells were cotransfected with luciferase vectors containing the c-myc 5′UTR and with Staufen1 or control plasmids. After transfection, the relative activity of luciferase was determined along with luciferase mRNA levels by qRT-PCR. Data are also normalized to luciferase levels in absence of Staufen1 and to an empty vector (n = 3). (G) Polysome profiling of proliferating stable C2C12 cells. Top, polysome profile obtained by continuous reading of absorbance at 254 nm. Bottom, levels of c-myc mRNAs were measured by qRT-PCR from 10 1-ml sucrose gradient fractions. (H) Cell proliferation assays performed with stable C2C12 cell lines (n = 3). Asterisks indicate significance (*p ≤ 0.05, **p ≤ 0.01).
Staufen1 is known to enhance the translation efficiency of a subpopulation of transcripts by binding to their 5′UTR (Dugre-Brisson et al., 2005). In addition, Staufen1 knockdown in U2OS cells reduces c-myc protein without affecting mRNA levels (Weidensdorfer et al., 2009). Therefore we examined whether Staufen1 overexpression promotes translation of c-myc mRNA. To test this, we first generated a luciferase construct containing the 5′UTR of c-myc. The 5′UTR-luciferase construct was then cotransfected with Staufen1-HA or control vectors. For these assays, HEK293T cells were used in order to reproduce conditions that led to the discovery of the translational role of Staufen1 (Dugre-Brisson et al., 2005). After transfection, expression of luciferase was monitored by a luciferase assay, and the level of luciferase mRNAs was measured by qRT-PCR. Overexpression of Staufen1-HA resulted in an increase in luciferase activity (Figure 8F). Under these conditions, luciferase transcript levels were in fact decreased, which, together with the increase in luciferase activity, strongly suggests that Staufen1 promotes translation of c-myc mRNAs (Figure 8F). In this context, it is important to note that a decrease in c-myc mRNA expression was also observed in Staufen1-HA myoblasts and after 48 h of differentiation (Figure 8C). This decrease in c-myc transcript level also occurs in a SMD-independent manner (Figure 7D).
To confirm whether overexpression of Staufen1-HA increases the translation of c-myc mRNA, we performed polysome fractionation experiments using proliferating stable cell lines. First, the optical density profiles of sucrose gradients were measured at 254 nm (Figure 8G). The top of the gradient contains free mRNAs and ribosomal subunits (40S, 60S, and 80S) whereas the bottom contains mRNAs associated with polysomes. Then we compared the expression profile of c-myc mRNAs along each gradient. As expected, qRT-PCR revealed enrichment in the levels of endogenous c-myc transcripts in polysomal fractions from Staufen1-HA–stable cells (clone #25) in comparison to controls (Figure 8G). To confirm this effect, C2C12 cells were transiently transfected with a Staufen1-HA construct and analyzed as described. First, we confirmed by Western blotting the Staufen1-HA overexpression (Supplemental Figure S1A). Then we determined by RT-PCR the distribution of c-myc mRNAs in the sucrose gradient and observed the same shift of c-myc mRNAs to the heavy polysome fraction upon Staufen1 overexpression (Supplemental Figure S1B). This shift in distribution of c-myc mRNAs to the polysome fractions shows that more ribosomes are loaded onto c-myc transcripts, thereby indicating that c-myc translation is enhanced by Staufen1-HA.
It is known that c-myc represses expression of genes involved in cell cycle, thereby inhibiting the irreversible proliferation arrest and promoting proliferation. Increased c-myc expression in Staufen1-HA–stable cells should therefore result in increased cell proliferation. To test this, we performed proliferation assays by measuring bromodeoxyuridine (BrdU) incorporation rate in control versus Staufen1-HA–stable cell lines. Briefly, cells were cultured in the presence of the thymidine analogue BrdU for 2 h. BrdU-positive cells were then visualized by immunofluorescence using anti-BrdU antibodies. We noticed a significant increase (p < 0.05) in cell proliferation in Staufen1-HA–stable cell lines (Figure 8H). Therefore the Staufen1-dependent increase in c-myc expression that we observed in terms of protein/mRNA steady-state measurements (Figure 8, A–C) and in c-myc translation efficiency (Figure 8, F and G) are both coherent with the dynamic change obtained in proliferation assays (Figure 8H). This increase in cell proliferation by Staufen1 observed after only 2 h of BrdU labeling reflects the relatively large effect of Staufen1 on the switch between proliferation and differentiation.
DISCUSSION
Regulation of Staufen1 and control of skeletal muscle cell development
Staufen1 has recently emerged as a multifunctional RNA-binding protein. Although a few studies have shown the importance of Staufen1 in skeletal muscle, little is known about its pattern of expression during embryonic and postnatal development. Furthermore, its precise role(s) in myogenic differentiation remain(s) fragmentary and conflicting (Belanger et al., 2003; Kim et al., 2007; Yamaguchi et al., 2008, 2012; Gong et al., 2009).
Here we found that expression of Staufen1 is high in E14.5 mouse muscle mass and that decreases progressively thereafter, becoming expressed at low levels in mature muscles. This pattern follows expression profiles observed during SkMC, HSMM, and mouse C2C12 differentiation and muscle degeneration/regeneration. These complementary findings are basically all in agreement, demonstrating that levels of Staufen1 are tightly regulated and decreased during muscle development and differentiation.
Given our convergent findings, we sought to prevent the decrease in Staufen1 expression during myogenesis. Our results show that sustained expression of Staufen1 clearly inhibits muscle differentiation by causing decreases in the expression of multiple key myogenic markers. In addition, our analyses revealed that Staufen1 overexpression increases c-myc protein levels without a parallel increase in mRNA levels, suggesting, in this case, that Staufen1 regulates the translational expression of c-myc. Moreover, down-regulation of Staufen1 by shRNAs decreases c-myc protein levels. These results are in agreement with previous findings showing that c-myc protein levels are significantly reduced upon Staufen1 knockdown in U2OS osteosarcoma cells, whereas c-myc mRNA remains unaffected (Weidensdorfer et al., 2009). In this context, it has been reported that Staufen1 has the ability to increase translation of specific mRNAs with structured 5′UTRs (Dugre-Brisson et al., 2005). Accordingly, here we show that Staufen1 overexpression increases translation of a luciferase reporter containing the 5′UTR of c-myc and relocates c-myc mRNAs to the polysomal fraction. Therefore it appears reasonable to argue that the increase in c-myc expression can be attributed to Staufen1 interacting with the c-myc 5′UTR to promote mRNA translation. Given the known role of c-myc in promoting cellular proliferation while inhibiting differentiation, our results uncover a new mechanism by which Staufen1 contributes to the balance between proliferation and differentiation and the regulation of muscle cell development.
We also demonstrated that the Staufen1-mediated inhibition of myogenesis cannot be rescued by expression of MyoD. This is consistent with the facts that 1) c-myc protein up-regulation is sufficient to inhibit differentiation of myogenic cells, and 2) the differentiation defect caused by c-myc cannot be bypassed by ectopic MyoD expression (Miner and Wold, 1991). Because increased rates of proliferation are incompatible with the induction of the differentiation program, we propose that Staufen1 overexpression affects the efficiency of myoblasts to differentiate into mature muscle fibers by at least partially promoting cell proliferation via this c-myc–dependent pathway.
Is Staufen1 promoting Staufen1-mediated decay during myogenic differentiation?
It is believed that Staufen1 controls mRNA stability by a mechanism called SMD. Binding of Staufen1 to a Staufen1-binding site (SBS) located in a 3′UTR triggers mRNA decay (Kim et al., 2005). This mechanism differs from NMD, as only Upf1, and not Upf2 or Upf3, is recruited to the target mRNA by Staufen1. SBS can be formed by intramolecular perfect base pairing forming a double-stranded stem-loop structure (Kim et al., 2007) or by intermolecular imperfect base pairing between Alu SINEs located in 3′UTRs of mRNAs and in long noncoding RNAs (Gong and Maquat, 2011; Wang et al., 2013).
It has been proposed that SMD and NMD are competitive pathways during myogenic differentiation (Gong et al., 2009). Although the levels of Staufen1 and Upf1 are decreased during myogenesis (Wang et al., 2013), the current model suggests that SMD efficiency increases whereas the effect of NMD decreases during myogenesis due to a reduced affinity of Upf2 for Upf1 (Gong et al., 2009). These patterns of Staufen1 expression and of its SMD partner Upf1 are thus opposite to the proposed effect of SMD during myogenic differentiation and appear physiologically counterintuitive. Moreover, Pax3 mRNA is a proposed SMD target decreased by Staufen1 in C2C12 myoblasts, thereby promoting myogenic differentiation (Gong et al., 2009). Pax3 is highly expressed in skeletal muscle progenitor cells, and its down-regulation is necessary for these cells to commit to the myoblast lineage (Lagha et al., 2008). However, and in agreement with other groups (Kuang et al., 2006; Collins et al., 2009; Kumar et al., 2009; Liu et al., 2010a), we were unable to detect Pax3 mRNA expression by qRT-PCR in proliferating C2C12 myoblasts (unpublished data). Therefore, whereas Pax3 down-regulation by SMD may constitute an event for muscle progenitor cell determination, it appears unlikely that it controls the myoblast to myotube transition.
In our study, we performed a candidate approach to identify muscle mRNA targets of Staufen1 involved in myoblast differentiation. Because expression of endogenous MyoD, MEF2A, and MEF2C mRNAs are decreased in stable C2C12 cells overexpressing Staufen1, we hypothesized that they may be SMD targets, despite the argument presented earlier. We used a strategy similar to the one used by the Maquat lab in the discovery of SMD by fusing candidate 3′UTRs downstream of a firefly luciferase reporter gene (Kim et al., 2007). None of the assessed targets decayed upon Staufen1 overexpression. We thus additionally tested the Arf1 3′UTR as a positive control for SMD. In our experiments, we used both human and mouse Arf1 3′UTRs, which contain two 19–base pair stem structures necessary to form the SBS (Kim et al., 2007). In contrast to previous work describing Arf1 as a canonical SMD target (Kim et al., 2005, 2007), our results failed to provide any evidence that Arf1 mRNA is destabilized by Staufen1 overexpression. While we were finalizing the manuscript for this article, two independent studies by the Moore and DesGroseillers labs using transcriptome-wide approaches also failed to obtain evidence of SMD upon Staufen1 overexpression on global mRNAs and on previously identified SMD targets, including Arf1 (Boulay et al., 2014; Ricci et al., 2014). These findings are in excellent agreement with ours and together raise questions as to the validity and functional significance of the current SMD model.
Impact for myotonic dystrophy
Myotonic dystrophy type 1 (DM1) is caused by the misregulation of RNA-binding proteins. In particular, CUGBP1 and Staufen1 are increased, and MBNL1 aggregates in the nucleus (Wheeler and Thornton, 2007; Lee and Cooper, 2009; O’Rourke and Swanson, 2009; Mahadevan, 2012; Ravel-Chapuis et al., 2012). Of interest, MBNL and CUGBP1 are linked to the posttranscriptional control of myogenic genes during skeletal muscle differentiation, which may alter the differentiation potential of DM1 muscle cells (Amack and Mahadevan, 2001; Amack et al., 2002; Timchenko et al., 2001; Squillace et al., 2002; Apponi et al., 2011). Given our present findings, it seems likely that expression of Staufen1 may also be altered in differentiating DM1 muscle, thereby further compromising differentiation of these cells. It appears timely to examine the pattern of expression of Staufen1 in developing DM1 muscle and to determine whether it contributes to pathogenic events as these muscle fibers differentiate and regenerate. Such information will provide new mechanistic insights into the complex muscle phenotype seen in DM1 patients.
MATERIALS AND METHODS
Constructs and antibodies
The constructs used were pcDNA3 (Invitrogen Life Technologies, Burlington, Canada), mStaufen1-HA3 and hStau155-HA3 (Wickham et al., 1999), and MyoD-Flag-myc expression vector (Liu et al., 2010b). The 3′UTR of human Arf1, mouse Arf1, MEF2A MEF2C, and c-myc was amplified by RT-PCR from C2C12 cells or mouse muscle extracts using primers containing a restriction site (underlined): hArf1 SBS (fwd-5′-ATTCTCGAGGTGAACGCGACCCCCCTCCCTCTCACTC-3′, rev-5′-CAGTCTAGACCAGGTGCCCATGGGCCTACATCCCC-3′); mArf1 (fwd-5′-ATTCTCGAGACCAGACCCCTCCCTCCC-3′, rev-5′-CAGTCTAGAAATAGTTAAGAGACTTTATTCTAA-3′) MyoD (fwd-5′-ATATTCTAGATCAGGTGCTTTGAGAGATCG-3′, rev-5′-ATATTCTAGATATAAATTAGCGTCTTTATTTCCAACA-3′); MEF2A (fwd-5′-ATATTCTAGAGGCTTCCTGGTTCATGTTTG-3′, rev-5′-ATATTCTAGATTCCAAGGTTCTGCTTGC-3′); MEF2C (fwd-5′-ATATTCTAGATCTGAAGGATGGGCAACAT-3′, rev-5′-ATATTCTAGAAAGAGATGCAGACCCAGATTT-3′); and c-myc (fwd-5′-TAAGCAGCTAGCTAAACTGACCTAACTCGAGGAGGAG-3′, rev-5′-TGCTTAGTCGACAGTTGGCCCAATTGTATTTTTTCCAATT-3′). PCR products were digested and subcloned into pmirGLO vector (Promega, Madison, WI) downstream of luciferase. The 5′UTR of c-myc was amplified by RT-PCR using the following primers: fwd-5′-ATATCTCGAGGATTGGGGTACGCGCTGC-3′ and rev-5′-ATATCTCGAGCGTCGTGGCTGTCTGCTG-3′. The PCR product was cloned upstream of luciferase into the modified pGL4.14 vector (Promega) containing a cytomegalovirus promoter. The orientation of inserts was determined by restriction digestion, and the integrity of sequences was confirmed by sequencing.
A Staufen1 shRNA construct was obtained by cloning the annealed complementary oligonucleotides 5′-GATCCCGGCAACGGTAACTGCCATGTTCAAGAGACATGGCAGTTACCGTTGCCTTTTTTCCAAA-3′ and 5′-AGCTTTTGGAAAAAAGGCAACGGTAACTGCCATGTCTCTTGAACATGGCAGTTACCGTTGCCGG-3′ into the BamHI–Hind III restriction sites of pRNAT-H1.1/Neo (Genscript, Piscataway, NJ) according to manufacturer's recommendations.
The antibodies used were anti-HA.11 (16B12; Covance, Montreal, Canada), anti-Staufen1, anti–c-myc and anti-GAPDH (AbCam, Toronto, Canada), anti-CUGBP1 and anti–β-actin (3B1 and C4, respectively; Santa Cruz Biotechnology, Santa Cruz, CA), anti-MyoD (MoAb 5.8A; BD Biosciences, Mississauga, Canada), anti-myogenin and anti-MyHC (F5D and MF20, respectively; Developmental Studies Hybridoma Bank, Iowa City, IA), and anti-laminin (Sigma-Aldrich, Oakville, Canada). Note that the band observed by Western blots for Staufen1 protein in mouse skeletal muscle cells is slightly above the 55-kDa mark. The band is specific for Staufen1, as its intensity is significantly decreased in protein extracts from cells expressing a shRNA targeting Staufen1 (see Figure 8, D and E).
Mouse muscle development and cardiotoxin injection
All surgical procedures were performed using aseptic conditions and were in complete agreement with the University of Ottawa Animal Care and Users Committee in compliance with the Guidelines of the Canadian Council on Animal Care and the Animals for Research Act. For developmental time course experiments, FVB/N mice were used. Skeletal muscles from mouse embryos were recovered at E14.5 and E18.5 and in newborns PN1. Given the small size of limbs at these ages, the whole muscle mass of the leg was used. TA muscles from 14-wk-old mice were used for adult samples. For degeneration/regeneration experiments, 25 μl of 10−5 M cardiotoxin (Latoxan, Rosans, France) were injected into TA muscles of 5- to 6-wk-old FVB/N mice to induce muscle degeneration and regeneration as described previously (Condrea, 1974; Clow and Jasmin, 2010). At different time points after injection, TA muscles were harvested, frozen in liquid nitrogen, and stored at −80°C until further analysis. Contralateral, saline-injected TA muscles were used as controls.
Cell culture, transfections, lentivirus infections, and stable cell lines
HeLa and HEK293T cells (American Type Culture Collection, Manassas, VA) were grown in growth medium (DMEM/10% fetal bovine serum [HyClone, Thermo Fisher Scientific, Ottawa, Canada], 100 U/ml penicillin, 100 μg/ml streptomycin). SkMCs and HSMM (Lonza, Allendale, NJ) were grown and differentiated according to manufacturer's recommendations. Mouse C2C12 cells (American Type Culture Collection) were maintained as myoblasts in growth medium as previously described (Ravel-Chapuis et al., 2007). To induce myogenic differentiation, cells were allowed to become confluent on Matrigel-coated (BD Biosciences) plates, and the medium was switched to differentiation medium (DMEM/2% horse serum [PAA Laboratories, Piscataway, NJ], 100 U/ml penicillin, 100 μg/ml streptomycin). Cell transfections were performed with 1 μg of DNA, Lipofectamine, and Plus Reagent (Invitrogen Life Technologies, Burlington, Canada) according to the manufacturer's instructions. Stable cell lines were obtained by selection of transfected cells with G418 (1 mg/ml; Life Technologies) until appearance of stably transfected clones. Clones expressing Staufen1-HA were screened by Western blotting, and they were maintained with G418 selection (0.5 mg/ml). Lentivirus production and cell transductions were performed as described (Ravel-Chapuis et al., 2012) using pLKO.1-shStau1 vectors (Open Biosystems GE Dharmacon, Lafayette, CO).
Immunofluorescence
Cells were fixed for 5 min in 1× phosphate-buffered saline (PBS) containing 1% formaldehyde. Cells were permeabilized for 5 min with 1× PBS and 0.5% Triton and blocked with 1× PBS with 1% bovine serum albumin (BSA). Cells were incubated with the primary antibody diluted in 1× PBS containing 1% BSA and 0.1% Triton for 1 h at 37°C or overnight at 4°C. Then, the cells were thoroughly washed with 1× PBS, and incubated for 1 h with Alexa secondary antibodies (Invitrogen Life Technologies). Slides were mounted with Vectashield mounting medium (Vector Labs, Burlington, Canada) containing 4′,6-diamidino-2-phenylindole (DAPI) for staining of nuclei. Fluorescence images were visualized by microscopy on a Z1 AxioImager upright microscope (Carl Zeiss, Toronto, Canada). Phase-contrast and low-magnification fluorescence images were obtained on an Axiovert S100 inverted microscope (Carl Zeiss). Images were processed with Photoshop CS5 (Adobe, San Jose, CA).
Differentiation index and fusion index
Myoblasts were seeded and allowed to differentiate for 24, 48, 72, or 96 h before fixation. Cells were immunostained for MyHC and DAPI as described, and images were acquired (10 random fields/condition). The fusion index (percentage of nuclei within myotubes having ≥3 nuclei) and differentiation index (percentage of nuclei within myotubes plus MyHC-positive mononucleated cells) were then determined.
Western blotting
Dissected muscle extracts were crushed in liquid nitrogen and muscle powder resuspended in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitors [Complete; Roche, Laval, Quebec]) or urea/thiourea buffer (7 M urea, 2 M thiourea, 65 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 100 mM dithiothreitol, 10 U of DNase I, and protease inhibitors [Complete]). Protein concentration was determined using the bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Ottawa, Canada) or CB-X Protein Assay kit (G-Bioscience, St. Louis, MO). Cells were washed and resuspended in RIPA buffer. Thirty micrograms of total proteins was separated by SDS–PAGE and transferred onto nitrocellulose membranes. Nonspecific binding was first blocked with 1× PBS containing 5% skim milk, and membranes were then incubated with primary antibodies. After thorough washing with 1× PBS with 0.05% Tween, membranes were incubated with horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). After several washes, signals were revealed using ECL reagents (PerkinElmer, Woodbridge, Canada) and autoradiographed with x-ray films (Thermo Fisher Scientific). Quantifications were performed with Image Lab software (Bio-Rad, Mississauga, Ontario).
RNA extraction, reverse transcription, and real-time quantitative PCR
Total RNA was extracted from samples using TRIzol (Invitrogen Life Technologies) or TriPure (Roche). One microgram of RNA was DNase treated (Ambion Life Technologies, Burlington, Canada), and cDNAs were synthesized using MuLV Reverse Transcriptase (Applied Biosystem Life Technologies). mRNA expression was evaluated by qRT-PCR (MX3005P; Stratagene, La Jolla, CA) using the QuantiTect SYBR Green PCR Kit (Qiagen, Toronto, Canada) according to the manufacturer's instructions. The sequences of the primers were as follows: MyoD (fwd-5′-ACTTTCTGGAGCCCTCCTGGC-3′, rev-5′-TTTGTTGCACTACACAGCATG-3′); MEF2A (fwd-5′-GAATGCCCAAAGGATAAGCA-3′, rev-5’-CAGCATTCCAGGGGAAGTAA-3′); MEF2C (fwd-5′-ATCTGCCCTCAGTCAGTTGG-3′, rev-5′-CAGCTGCTCAAGCTGTCAAC-3′); myogenin (fwd-5′-CTACAGGCCTTGCTCAGCTC-3′, rev-5′-AGATTGTGGGCGTCTGTACG-3′); and c-myc (fwd-5′-GCCCAGTGAGGATATCTGGA-3′, rev-5′-ATCGCAGATGAAGCTCTGGT-3′). The measures were normalized to cyclophylin-B levels (fwd-5′-GATGGCACAGGAGGAAAGAG-3′, rev-5′-AACTTTGCCGAAAACCACAT-3′). For luciferase experiments, qRT-PCR was performed with luciferase (fwd-5′-TGCAAAAGATCCTCAACGTG-3′, rev-5′-AATGGGAAGTCACGAAGGTG-3′) and normalized to human 18S (fwd-5′-GTAACCCGTTGAACCCCATT-3′, rev-5′-CCATCCAATCGGTAGTAGCG-3′).
Luciferase reporter assay
After transfection, assays for luciferase enzymatic activity were performed on cell lysates using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Measures were performed using a luminometer (Lumat LB 9507; Berthold Technologies, Oak Ridge, TN).
Proliferation assay
Cells were seeded on glass coverslips and left to proliferate overnight. Proliferation was assessed using the BrdU Labeling and Detection Kit I (Roche) according to manufacturer's instructions, using a 2-h BrdU incubation time. Coverslips were mounted with Vectashield mounting medium (Vector Labs) containing DAPI for staining of nuclei and visualized as described.
Polysome fractionation experiments
The 50% confluent proliferating stable cell lines were grown on 150-mm plates. Cells were treated with 0.1 mg/ml CHX (Sigma-Aldrich) for 5 min in fresh DMEM. Cells were washed twice with PBS containing CHX and subsequently lysed in RNA lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris, pH 7.4, 1% Triton X-100, 0.1 mg/ml CHX, 100 U/ml RNasin). Nuclei and cellular debris were removed by centrifugation steps (3000 rpm for 5 min at 4°C and 14,000 rpm for 5 min at 4°C). Five hundred microliters of the lysates was layered on continuous sucrose gradients (10–50% sucrose in 15 mM MgCl2, 15 mM Tris, pH 7.4, 0.3 M NaCl). Centrifugations were carried out at 39,000 rpm in a SW41-Ti rotor at 4°C for 90 min. Then 1-ml fractions were collected at a flow rate of 1 ml/min from top to bottom of the gradient while absorbance was measured continuously at 254 nm. Samples were digested with proteinase K, and total RNA was extracted.
Statistical analysis
Student's t tests were used to determine whether differences between groups were significant. The level of significance was set at p ≤ 0.05. *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001. Means ± SEM are presented throughout, unless otherwise specified.
Supplementary Material
Acknowledgments
We thank our colleagues for helpful discussion throughout the course of this work. We thank A. Blais for providing us with the MyoD expression vector, J. Lunde for technical assistance with cardiotoxin injection experiments, and J. Langill for help with lentivirus production and cell transductions. This work was supported by the Canadian Institutes of Health Research, the Rachel Fund (Canadian Institutes of Health Research/Institute of Musculoskeletal Health and Arthritis, and Muscular Dystrophy Canada), the Association Française contre les Myopathies, and the Muscular Dystrophy Association (United States). A.R.-C. was the recipient of a Postdoctoral Fellowship from the Association Française contre les Myopathies and from the Canadian Institutes of Health Research during the course of this work and also benefited from a Development Grant from the Muscular Dystrophy Association.
Abbreviations used:
- ARE
AU-rich elements
- BrdU
bromodeoxyuridine
- CTX
cardiotoxin
- CUGBP
CUG-binding protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA
hemagglutinin
- HuR
Hu antigen R
- KSRP
K-homology splicing regulator protein
- MBNL
muscleblind-like protein
- MEF2
myocyte enhancer factor-2
- MyHC
myosin heavy chain
- NMD
nonsense-mediated mRNA decay
- Pax3
paired box 3
- qRT-PCR
quantitative reverse transcription-PCR
- SBS
Staufen1-binding site
- shRNA
short hairpin RNA
- SINEs
short-interspersed elements
- SMD
Staufen1-mediated mRNA decay
- TA
tibialis anterior
- UTR
untranslated region
Footnotes
A.R.-C., T.E.C., M.-L.B.-C., G.B., and C.T.R. performed the experiments. A.R.-C., T.E.C., and B.J.J. designed the experiments, analyzed the data, and wrote the manuscript.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-04-0895) on September 10, 2014.
REFERENCES
- Amack JD, Mahadevan MS. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum Mol Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- Amack JD, Reagan SR, Mahadevan MS. Mutant DMPK 3’-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD. J Cell Biol. 2002;159:419–429. doi: 10.1083/jcb.200206020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirouche A, Tadesse H, Lunde JA, Belanger G, Cote J, Jasmin BJ. Activation of p38 signaling increases utrophin A expression in skeletal muscle via the RNA-binding protein KSRP and inhibition of AU-rich element-mediated mRNA decay: implications for novel DMD therapeutics. Hum Mol Genet. 2013;22:3093–3111. doi: 10.1093/hmg/ddt165. [DOI] [PubMed] [Google Scholar]
- Apponi LH, Corbett AH, Pavlath GK. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol Sci. 2011;32:652–658. doi: 10.1016/j.tips.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp P, Nassif C, Hillock S, van der Giessen K, von Roretz C, Jasmin BJ, Gallouzi IE. The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation. Cell Death Differ. 2010;17:1588–1599. doi: 10.1038/cdd.2010.34. [DOI] [PubMed] [Google Scholar]
- Belanger G, Stocksley MA, Vandromme M, Schaeffer L, Furic L, DesGroseillers L, Jasmin BJ. Localization of the RNA-binding proteins Staufen1 and Staufen2 at the mammalian neuromuscular junction. J Neurochem. 2003;86:669–677. doi: 10.1046/j.1471-4159.2003.01883.x. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4, a008342 doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bismuth K, Relaix F. Genetic regulation of skeletal muscle development. Exp Cell Res. 2010;316:3081–3086. doi: 10.1016/j.yexcr.2010.08.018. [DOI] [PubMed] [Google Scholar]
- Boulay K, Ghram M, Viranaicken W, Trepanier V, Mollet S, Frechina C, DesGroseillers L. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res. 2014;42:7867–7883. doi: 10.1093/nar/gku506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel C, Rehbein M, Kreienkamp HJ, Buck F, Richter D, Kindler S. Characterization of Staufen 1 ribonucleoprotein complexes. Biochem J. 2004;384:239–246. doi: 10.1042/BJ20040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Vincent SD. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr Opin Genet Dev. 2009;19:444–453. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Clow C, Jasmin BJ. Brain-derived neurotrophic factor regulates satellite cell differentiation and skeletal muscle regeneration. Mol Biol Cell. 2010;21:2182–2190. doi: 10.1091/mbc.E10-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Gnocchi VF, White RB, Boldrin L, Perez-Ruiz A, Relaix F, Morgan JE, Zammit PS. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS One. 2009;4:e4475. doi: 10.1371/journal.pone.0004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrea E. Membrane-active polypeptides from snake venom: cardiotoxins and haemocytotoxins. Experientia. 1974;30:121–129. doi: 10.1007/BF01927688. [DOI] [PubMed] [Google Scholar]
- Deschenes-Furry J, Angus LM, Belanger G, Mwanjewe J, Jasmin BJ. Role of ELAV-like RNA-binding proteins HuD and HuR in the post-transcriptional regulation of acetylcholinesterase in neurons and skeletal muscle cells. Chem Biol Interact. 2005;157-158:43–49. doi: 10.1016/j.cbi.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5’ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftimie R, Brenner HR, Buonanno A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc Natl Acad Sci USA. 1991;88:1349–1353. doi: 10.1073/pnas.88.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina NH, Hausburg M, Betta ND, Pulliam C, Srivastava D, Cornelison D, Olwin BB. A role for RNA post-transcriptional regulation in satellite cell activation. Skelet Muscle. 2012;2:21. doi: 10.1186/2044-5040-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiol A, St Johnston D. Staufen targets coracle mRNA to Drosophila neuromuscular junctions and regulates GluRIIA synaptic accumulation and bouton number. Dev Biol. 2014;392:153–167. doi: 10.1016/j.ydbio.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell. 2009;20:3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Lagha M, Sato T, Bajard L, Daubas P, Esner M, Montarras D, Relaix F, Buckingham M. Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7. Cold Spring Harb Symp Quant Biol. 2008;73:307–315. doi: 10.1101/sqb.2008.73.006. [DOI] [PubMed] [Google Scholar]
- Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, Morris QD, Lipshitz HD. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res. 2013;41:9438–9460. doi: 10.1093/nar/gkt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Liu C, Gersch RP, Hawke TJ, Hadjiargyrou M. Silencing of Mustn1 inhibits myogenic fusion and differentiation. Am J Physiol Cell Physiol. 2010a;298:C1100–1108. doi: 10.1152/ajpcell.00553.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 2010b;38:6857–6871. doi: 10.1093/nar/gkq585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan MS. Myotonic dystrophy: is a narrow focus obscuring the rest of the field. Curr Opin Neurol. 2012;25:609–613. doi: 10.1097/WCO.0b013e328357b0d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Fortes P, Beloso A, Dotti C, Ortin J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev MP, Ravichandran M, Khan MF, Schriemer DC, Mouland AJ. Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1. Front Microbiol. 2012;3:367. doi: 10.3389/fmicb.2012.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Wold BJ. c-myc inhibition of MyoD and myogenin-initiated myogenic differentiation. Mol Cell Biol. 1991;11:2842–2851. doi: 10.1128/mcb.11.5.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Orengo JP, Ward AJ, Cooper TA. Alternative splicing dysregulation secondary to skeletal muscle regeneration. Ann Neurol. 2011;69:681–690. doi: 10.1002/ana.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J Biol Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Belanger G, Yadava RS, Mahadevan MS, Desgroseillers L, Cote J, Jasmin BJ. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J Cell Biol. 2012;196:699–712. doi: 10.1083/jcb.201108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Vandromme M, Thomas JL, Schaeffer L. Postsynaptic chromatin is under neural control at the neuromuscular junction. EMBO J. 2007;26:1117–1128. doi: 10.1038/sj.emboj.7601572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol. 2014;21:26–35. doi: 10.1038/nsmb.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squillace RM, Chenault DM, Wang EH. Inhibition of muscle differentiation by the novel muscleblind-related protein CHCR. Dev Biol. 2002;250:218–230. doi: 10.1006/dbio.2002.0798. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tubing F, et al. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci USA. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Roretz C, Beauchamp P, Di Marco S, Gallouzi IE. HuR and myogenesis: being in the right place at the right time. Biochim Biophys Acta. 2011;1813:1663–1667. doi: 10.1016/j.bbamcr.2011.01.036. [DOI] [PubMed] [Google Scholar]
- Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Naiki T, Irie K. Stau1 regulates Dvl2 expression during myoblast differentiation. Biochem Biophys Res Commun. 2012;417:427–432. doi: 10.1016/j.bbrc.2011.11.133. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Oohinata R, Naiki T, Irie K. Stau1 negatively regulates myogenic differentiation in C2C12 cells. Genes Cells. 2008;13:583–592. doi: 10.1111/j.1365-2443.2008.01189.x. [DOI] [PubMed] [Google Scholar]
- Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.