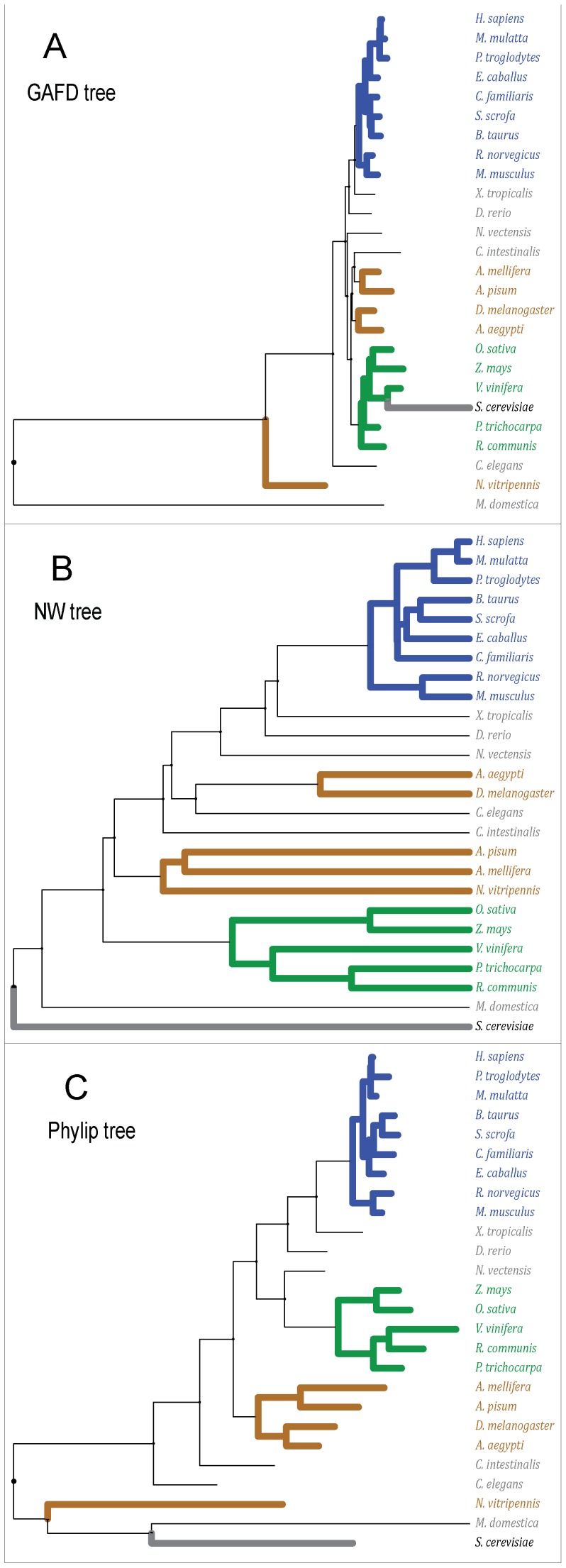
Figure 3. Depiction of phylogenetic trees for the ribosomal 18S subunit gene of 26 selected species.
(A and B) Trees computed with GAFD and NW, respectively. (C) Maximum parsimony-bootstrapped Phylip tree. The species assessed and their corresponding KEGG entries are: Acyrthosiphon pisum (api:100145839), Aedes aegypti (aag:AaeL_AAEL009747), Apis mellifera (ame:552726), Bos taurus (bta:326602), Caenorrhabditis elegans (cel:Y57G11C.16), Canis familiaris (cfa:403685), Ciona intestinalis (cin:100182116), Danio rerio (dre:192300), Drosophila melanogaster (dme:Dmel_CG8900), Equus caballus (ecb:100052654), Homo sapiens (hsa:6222), Macaca mulatta (mcc:713939), Monodelphis domestica (mdo:100027117), Mus musculus (mmu:20084), Nasonia vitripennis (nvi:100117049), Nematostella vectensis (nve:NEMVE_v1g245261), Oryza sativa (osa:4334407), Pan troglodytes (ptr:455055), Populus trichocarpa (pop:POPTR_551159), Rattus norvegicus (rno:100360679), Ricinus communis (rcu:RCOM_0557270), Saccharomyces cerevisiae (sce:YDR450W), Sus scrofa (ssc:396980), Vitis vinifera (vvi:100245272), Xenopus tropicalis (xla:414719), Zea mays (zma:100285246). The trees are color-coded for the relevant phylogenetic groups, namely blue for eutherian mammals, green for plants and brown for insects. S. cerevisiae is bolded as reference.