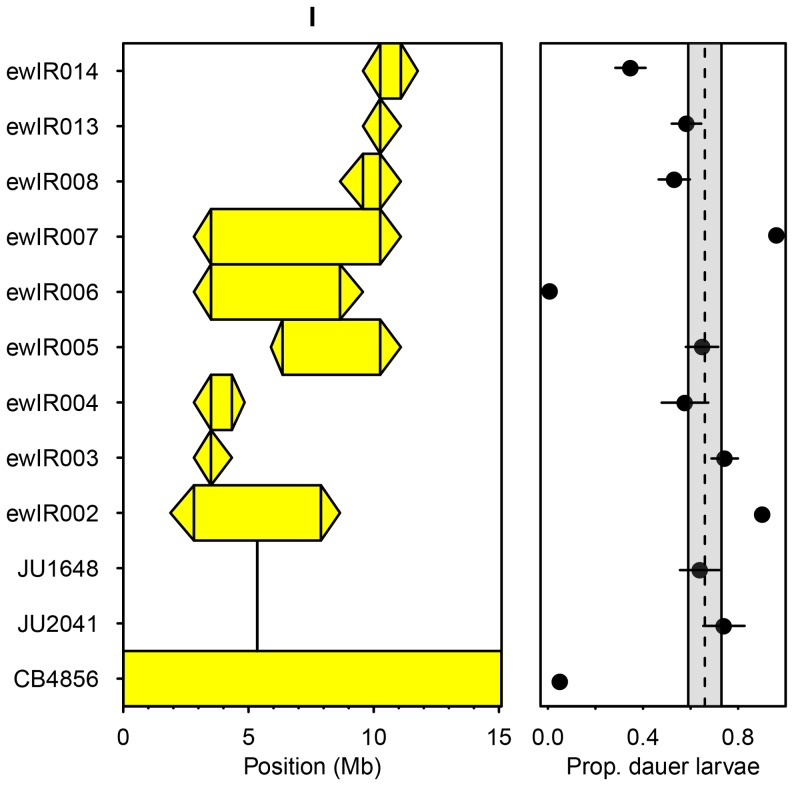
Figure 5. Variation at nath-10 does not affect dauer larvae formation in response to defined amounts of pheromone.
Reanalysis of ILs with introgressions in the region of nath-10 and of JU1648 and JU2041, lines with very fine introgressions of nath-10(haw6805). The nath-10(haw6805) allele is the only common difference distinguishing JU2041 and JU1648 from N2, and JU2041 contains only two known alleles that differ from N2, nath-10(haw6805) and mfP19 [19]. The CB4856 introgression per IL is shown by the colored rectangle and triangles joining adjacent CB4856 and N2 markers (left hand panel), and the proportion of dauer larvae that developed in each line (right hand panel) in dauer larvae formation assays with 40 µl of dauer pheromone extract and 20 µl of 1% w/v E. coli. Error bars represent ±1 S.E. and the dashed lines and shaded bar represent the mean ±1 S.E. of N2.