Abstract
Th1 and Th2 cell fates are traditionally viewed as mutually exclusive, but recent work suggests that these lineages may be more plastic than previously thought. When isolating splenic CD4+ T cells from mice infected with the parasitic helminth Schistosoma mansoni, we observed a defined population of IFN-γ/IL-4 double-positive cells. These IFN-γ+IL-4+ cells showed differences in DNA methylation at the Ifng and Il4 loci when compared with IFN-γ+IL-4− (Th1) and IFN-γ−IL-4+ (Th2) cells, demonstrating that they represent a distinct effector cell population. IFN-γ+IL-4+ cells also displayed a discrete DNA methylation pattern at a CpG island within the body of the Gata3 gene, which encodes the master regulator of Th2 identity. DNA methylation at this region correlated with decreased Gata3 levels, suggesting a possible role in controlling Gata3 expression. These data provide important insight into the molecular mechanisms behind the co-existence of Th1 and Th2 characteristics.
Keywords: DNA methylation, Gata3, Helminth, Th1/Th2
Introduction
Th1 and Th2 cells are specialized subsets of CD4+ T helper cells that respond to different modes of infection and which express the signature cytokines IFN-γ and IL-4, respectively. Transcription factors are involved in specifying Th1 and Th2 cell fate: T-bet is responsible for Th1 lineage determination while Gata3 is the key factor involved in Th2 development 1,2. Although Th1 and Th2 are generally thought of as mutually exclusive cell fates, since factors made by one cell type antagonize the generation of the other 1,3, several studies suggest some flexibility in Th1 and Th2 cell identity. Th1 and Th2 cytokines can be co-expressed by T-cell clones in vitro 4,5, and in vitro generated Th2 cells can be reprogrammed by Th1-promoting lymphocytic choriomeningitis virus to adopt a Gata3+T-bet+ phenotype 6. Similarly, in vitro derived Th1 cells can be converted to IL-4-producing Th2 cells 7, and T cells with both Th1 and Th2 characteristics can be found during helminth infection 8,9. Importantly, the molecular details of how CD4+ T cells can stably maintain both Th1 and Th2 characteristics remain unclear.
Epigenetic mechanisms, including DNA methylation occurring at the dinucleotide sequence CpG, can be involved in maintaining Th1 and Th2 identity 10,11. When DNA methylation occurs at gene promoters, it is associated with transcriptional silencing 12,13. In general, the Ifng locus shows reduced DNA methylation upon Th1 differentiation, while the Il4/Il5/Il13 locus loses DNA methylation upon Th2 differentiation. This is consistent with upregulation of Ifng expression during Th1 differentiation and induction of Il4 and other Th2 cytokines during Th2 differentiation 10,11.
In vertebrates, the genome is punctuated by CpG islands (CGIs), which have an increased density of CpG dinucleotides compared to the rest of the genome and an elevated G+C base composition 14. Although CGIs are usually unmethylated, DNA methylation can occur during normal development 13. CGIs frequently associate with gene promoters, although they also occur within and between annotated genes 15. We recently carried out a genome-wide survey of DNA methylation at CGIs in immune cells and identified just one CGI methylation difference between Th1 and Th2 cells differentiated in vitro. This occurred at a CGI within the body of the gene encoding Gata3, the master regulator of Th2 cell identity 16.
We wanted to investigate DNA methylation of Ifng and Il4 in a physiologically relevant infection setting. As Gata3 regulates Th2 differentiation, we isolated CD4+ T cells from mice infected with the Th2-inducing parasitic helminth Schistosoma mansoni 17. We observed splenocytes positive for both IFN-γ and IL-4 in infected mice, in addition to conventional Th1 and Th2 cells producing only IFN-γ or IL-4. The IFN-γ+IL-4+ cells displayed a distinct DNA methylation signature at key cytokine genes and at Gata3, suggesting that methylation patterns may be important for allowing the stable co-existence of Th1 and Th2 phenotypes. Our data further challenge the paradigm that Th1 and Th2 cell fates are mutually exclusive by demonstrating that IFN-γ+IL-4+ cells are a discrete cell population, different from Th1 and Th2 cells with respect to DNA methylation as well as cytokine production. Our data also suggest a potential role for the Gata3 CGI in regulating Gata3 expression and highlight possible regulatory significance for intragenic CGI methylation more generally.
Results and discussion
IFN-γ+IL-4+ cells are generated during S. mansoni infection
In order to examine DNA methylation in an in vivo infection setting we isolated splenic CD4+ T cells from mice that had been infected with S. mansoni for 8 weeks and from age-matched uninfected controls (Fig.1A). A marked proportion of CD4+ T cells displayed properties of both Th1 and Th2 cells in that they simultaneously made both IFN-γ and IL-4 8 (Fig.1B and Supporting Information Fig. 1). Conventional IFN-γ+IL-4− Th1 cells and IFN-γ−IL-4+ Th2 cells were also present, consistent with previous reports 18,19 and CD4+ T cells from uninfected mice showed significantly less expression of IFN-γ or IL-4 (Fig.1B). IFN-γ+IL-4+ cells were observed in five separate S. mansoni infections with the proportion varying from approximately 2–9% of CD4+ T cells (data not shown), demonstrating that IFN-γ+IL-4+ cells can be found in the spleen in a Th2-dominated infection setting.
Figure 1.
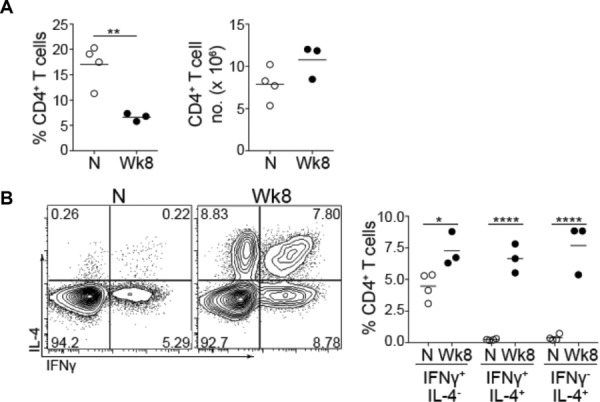
IFN-γ+IL-4+ cells are generated during Schistosoma mansoni infection. Splenocytes were isolated from infected mice taken from the same experiment (Wk 8) or uninfected age-matched controls (N). (A) The proportion and number of splenic CD4+ T cells was assessed by flow cytometry. Each symbol represents an individual animal and the mean of each sample group is shown as a horizontal line. Statistical significance was assessed using a Student's t-test and data are representative of two independent experiments. (B) Purified splenic CD4+ T cells were stimulated with PMA, ionomycin, and GolgiStop. FACS plots show intracellular staining of IFN-γ and IL-4. Dead cells, doublets and TCR-β−CD4− cells were excluded prior to assessing cytokine expression (Supporting Information Fig. 1). Plots are representative of five independent infections where either pools of spleens were analyzed or animals were analyzed separately. Results were similar in both cases. The percentage of splenic CD4+ cells expressing IFN-γ alone, both IFN-γ and IL-4, or IL-4 alone for individual mice taken from the same experiment are plotted on the right hand side of the figure, significance was assessed using two-way ANOVA. *p < 0.05, **p < 0.01 and ****p < 0.0001.
A balance between Th1 and Th2 responses is critical for host survival in S. mansoni infection 17. The Th2 response is crucial for limiting disease in the early stages of the infection 20, while prolonged or excessive Th2 responses lead to liver fibrosis and decreased survival, mediated predominantly by IL-13 21. IFN-γ may help to counter-regulate such Th2-mediated fibrotic disease during infection 22–24. Thus, IFN-γ+IL-4+ double positive cells may help maintain a balance between extreme Th1 and Th2 polarization during S. mansoni infection.
IFN-γ+IL-4+ cells show a distinct DNA methylation pattern at cytokine gene loci and Gata3
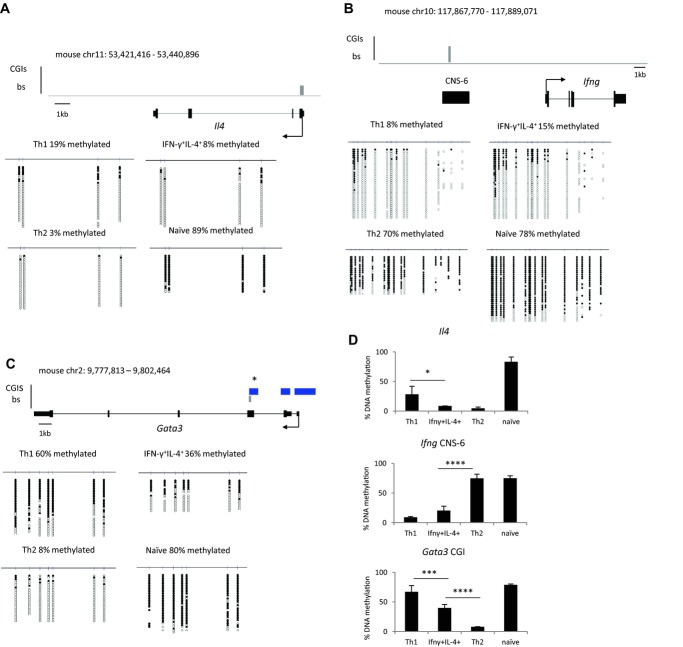
To examine whether IFN-γ+IL-4+ cells are distinct on a molecular level from Th1 and Th2 cells, we examined DNA methylation of key immune system genes. IFN-γ+IL-4− (Th1), IFN-γ−IL-4+ (Th2), and IFN-γ+IL-4+ cell populations were FACS sorted, with purities of 96.8–98% (Supporting Information Fig. 2), prior to methylation analysis by bisulfite genomic sequencing. DNA from the isolated cells was treated with sodium bisulfite, which converts unmethylated cytosines to uracil but leaves methylated cytosines unconverted, then PCR was performed to amplify genomic regions of interest, which were subsequently sequenced 25. First we examined DNA methylation at the key cytokine gene loci, Ifng and Il4, which have been reported to differ in methylation status between in vitro generated Th1 and Th2 cells 10. This revealed a distinct DNA methylation signature for IFN-γ+IL-4+ cells compared with conventional Th1 and Th2 cells. In double positive cells, both the Il4 promoter and the Ifng CNS-6 regulatory region showed significant demethylation (Fig.2A and B). Conventional Th1 and Th2 cells lacked methylation at the locus for their signature cytokine while the locus for the opposing cytokine was more extensively methylated. In CD4+ cells isolated from uninfected mice both Il4 and Ifng were completely methylated (Fig.2A and B). DNA methylation is frequently associated with gene repression and these results are broadly consistent with the fact that Th1 cells do not express Il4, Th2 cells do not express Ifng, while IFN-γ+IL-4+ cells, which have low levels of DNA methylation at Il4 and Ifng, express both genes. However, it is worth noting that in Th1 cells the Il4 promoter showed a dramatic decrease in DNA methylation compared with naïve controls (Fig.2A). This could suggest that demethylation of the Il4 locus is a general feature of CD4+ T cells in Th2 environments. Nevertheless, our data demonstrate that ex vivo IFN-γ+IL-4+, Th1, and Th2 cells are distinct from each other with respect to DNA methylation as well as cytokine production. During S. mansoni infection, the spleen is an accepted site for assessing responding lymphocytes, which include circulating effector and effector/memory CD4+ T cells 26. An important next step in our studies will be to assess the methylation signature of IFN-γ+IL-4+ T cells isolated from effector sites such as the liver.
Figure 2.
IFN-γ+IL-4+ cells show a distinct DNA methylation signature at Il4, Ifng and Gata3. (A) Il4 promoter, (B) Ifng CNS-6, (C) Gata3 gene body CGI (*) DNA methylation was determined using bisulfite sequencing. The relevant gene and the region analyzed by bisulfite (grey bar) are shown (upper panel). The arrow indicates the origin and direction of transcription and, where present, CGIs are represented by blue bars. Bisulfite sequencing data for Th1, Th2, IFN-γ+IL-4+ CD4+ T cells isolated from infected mice and naïve CD4+ T cells isolated from uninfected controls are also shown (lower panel). Filled circles represent methylated CpG residues, empty circles represent unmethylated CpGs and each row corresponds to an individual sequenced clone. (A–C) Data shown are from single experiments representative of two independent experiments performed. (D) The percentage DNA methylation is shown as mean + SD of two independent experiments. Statistical significance of methylation differences between Th1, Th2, and IFN-γ+IL-4+ cells was assessed using QUMA software and Mann–Whitney U test, * p < 0.05, *** p < 0.001 and **** p < 0.0001.
We have previously shown that the only CGI methylation difference between in vitro differentiated Th1 and Th2 cells occurs at a CGI in the body of the Gata3 gene, overlapping its third exon (Fig.2C) 16. We assessed DNA methylation of this region in the different CD4+ T-cell populations isolated ex vivo from S. mansoni infection. IL-4-producing Th2 cells showed almost complete demethylation of the Gata3 CGI (8% methylation). In contrast, Th1 cells were heavily methylated at this CGI (60% methylation). IFN-γ+IL-4+ cells had Gata3 DNA methylation levels intermediate between those of Th1 and Th2 cells (36% methylation, Fig.2C). Examination of a different bisulfite PCR amplicon within the Gata3 gene body CGI yielded similar results (Supporting Information Fig. 3A and B). For all regions interrogated, two independent experiments gave comparable DNA methylation levels in each of the cell populations (Fig.2D).
The distinct DNA methylation patterns observed at key immune gene loci in IFN-γ+IL-4+ cells demonstrate that these cells are different on a molecular level from Th1 and Th2 cells and therefore represent a distinct CD4+ T cell population generated during S. mansoni infection. Furthermore, this unique DNA methylation signature may represent an epigenetic state that allows for inherent flexibility in the identity of these double positive cells.
Methylation of the Gata3 gene body CGI negatively correlates with gene expression
It is well established that DNA methylation of a promoter CGI correlates with silencing of the associated gene 12,13, but the effect of gene body CGI methylation on transcription is less clear. We assessed Gata3 expression using intracellular staining and FACS in Th1, Th2, and IFN-γ+IL-4+ cells and compared Gata3 levels to DNA methylation levels within each population (Fig.3A and B). Methylation of the Gata3 intragenic CGI correlated with decreased expression of Gata3, despite the fact that this CGI is not located at the gene's promoter (Fig.3B). For example, in Th1 cells where this CGI is heavily methylated, only 7% of cells stained positive for Gata3. In contrast, in Th2 cells where Gata3 is demethylated, 80% of cells expressed Gata3. IFN-γ+IL-4+ cells showed Gata3 expression intermediate between Th1 and Th2 cells, consistent with their intermediate DNA methylation levels (Fig.3A and B). Moderate Gata3 expression and gene body methylation levels in IFN-γ+IL-4+ cells may be important for allowing them to possess both Th1 and Th2 characteristics.
Figure 3.
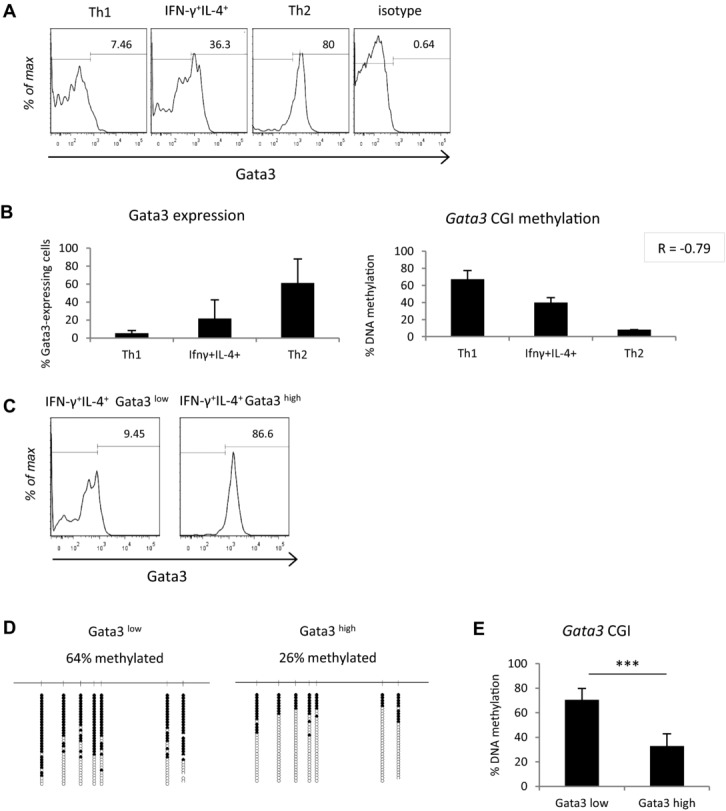
Gata3 gene body CGI methylation negatively correlates with gene expression. (A) Gata3 expression as assessed by flow cytometry in Th1, Th2, and IFN-γ+IL-4+ cells. (B) The percentage of Gata3-expressing cells in spleens isolated from S. mansoni infection is plotted alongside the overall percentage DNA methylation, as assessed by bisulfite sequencing, at the Gata3 CGI for each cell population. Pearson's correlation coefficient (R) for the two datasets is shown in top right hand corner. (C) Sorting of IFN-γ+IL-4+ cells into Gata3low and Gata3high cells by FACS. (D) Gata3 methylation of Gata3low and Gata3high cells as quantified by bisulfite sequencing, labelling as in Fig.2. (E) Mean Gata3 gene body CGI methylation levels in Gata3low and Gata3high cells. Statistical significance was assessed using QUMA software and Mann–Whitney U test, ***p < 0.001. (A, C, D) Data shown are from a single experiment, representative of two independent experiments performed. (B, E) Data are shown as mean + SD of two independent experiments. FACS experiments are gated on FSC-A and SSC-A, FSC-A and FSC-W, CD4 expression, and lastly on IFN-γ and IL-4 production.
We then examined the relationship between Gata3 methylation and gene expression more closely within the IFN-γ+IL-4+ population. IFN-γ+IL-4+ cells were FACS sorted into two additional populations: those that expressed high levels of Gata3 and those that expressed low levels (Fig.3C and Supporting Information Fig. 4A). DNA methylation at the Gata3 intragenic CGI was then assessed by bisulfite sequencing. Gata3low cells displayed a significantly higher overall level of DNA methylation than Gata3high cells (64% compared with 26%, p ≤ 0.0005) with approximately half of the DNA molecules in the Gata3low cells methylated at all sites tested (Fig.3D and E). This is consistent with an association between gene body CGI methylation and transcriptional repression in an infection setting. Further, as expected, Gata3high cells showed moderately higher levels of IL-4 expression, and lower levels of IFN-γ expression, when compared with Gata3low cells (Supporting Information Fig. 4B).
Gata3 possesses a promoter CGI in addition to its gene body CGI (Fig.2C) 16. Notably, DNA methylation analysis of the promoter CGI showed that it remained unmethylated in all cell populations despite differences in Gata3 expression (Supporting Information Fig. 3C). This finding highlights a potential role for the Gata3 gene body CGI rather than the promoter CGI in regulating Gata3 gene expression via DNA methylation. It is possible that the gene body CGI is an alternative promoter for Gata3, or acts as another kind of regulatory element, such as an enhancer, that functions only when it is demethylated. Another explanation could be that gene body CGI methylation inhibits transcriptional elongation and therefore its removal facilitates Gata3 expression 27.
Tbx21, the locus from which T-bet is expressed, has a promoter CGI but no gene body CGI 15. Analysis of existing DNA methylation data for naïve CD4+ cells and in vitro differentiated Th1 and Th2 cells showed that the Tbx21 promoter CGI is completely unmethylated in all of these cell types (16, and data not shown). Based on this, as well as the lack of methylation at the Gata3 promoter (Supporting Information Fig. 3C), we would suggest that it is unlikely that the DNA methylation status of Tbx21 changes in CD4+ T cells from S. mansoni infection. However, it is possible that expression of T-bet along with expression of Gata3 contributes to the dual identity of IFN-γ+IL-4+ cells, even if it is not associated with differences in DNA methylation.
The nature of the regulatory role played by the Gata3 gene body CGI and whether DNA demethylation of this CGI is a cause or consequence of Gata3 expression warrants further investigation. Irrespective, our results indicate that intragenic CGI methylation might be more reflective of transcriptional activity generally than DNA methylation at promoter CGIs, as these are almost always unmethylated 15,28.
Concluding remarks
In summary, we have identified a population of splenic CD4+ T cells generated during S. mansoni infection, which expresses both IFN-γ and IL-4 simultaneously. These IFN-γ+IL-4+cells are molecularly distinct from Th1 and Th2 cells as they show a unique DNA methylation signature at key immune genes, suggesting that DNA methylation is important for allowing co-existence of Th1 and Th2 characteristics. We have also uncovered a relationship between methylation of a CGI in the body of Gata3 and repression of Gata3 expression. This raises the interesting possibility that intragenic CGI methylation may represent a novel general mechanism of gene regulation in immune cells.
Materials and methods
Animals and S. mansoni infection
Experiments were performed using female C57BL/6 mice, which were maintained under specific pathogen-free conditions and used at 8–12 weeks of age. Experiments were conducted under a Project License granted by the Home Office (United Kingdom) in accordance with local guidelines. Mice were infected percutaneously with 40–80 S. mansoni cercariae, as previously described 29. Infections were allowed to proceed for 8 weeks prior to culling and spleen cell isolation.
T-cell isolation, culture, and FACS
CD4+ T cells were positively or negatively selected from naïve and infected spleens (Miltenyi Biotec or Life Technologies) and cultured in X-Vivo 15 medium (Lonza, BioWhittaker) supplemented with l-glutamine and 2-mercaptoethanol, and then stimulated with PMA (10 ng/mL), ionomycin (1 μg/mL), and Golgistop (BD Biosciences; 1:1000) for 4–5 hours at 37°C, 5% CO2. Cells were fixed, permeabilized and stained with antibodies against IFN-γ (Biolegend), IL-4 (BD Biosciences), and Gata-3 (BD Biosciences). For DNA methylation experiments, 6–10 spleens were pooled in order to obtain adequate numbers of cells for downstream analysis. Sorting was carried out using a BD FACS Aria. For statistical analysis, GraphPad Prism software was used to perform Student's t-test or ANOVA where appropriate.
Bisulfite genomic sequencing
DNA was isolated from sorted cells according to standard protocols. Bisulfite treatment and sequencing was performed as previously described 30. Statistical analysis was carried out using the QUMA package 31 and statistical significance was assessed by Mann–Whitney U test. Sequences of primers used for PCR amplification of bisulfite treated DNA are available upon request.
Acknowledgments
We thank Martin Waterfall for FACS sorting, Lucy Jones for help with S. mansoni infections, and Heather Owen for comments on the manuscript. We also thank Richard Preziosi for advice on statistical analysis. This work was funded by a Wellcome Trust PhD studentship to AD and grants to AB and ASM from the MRC (United Kingdom) and the Wellcome Trust.
Glossary
- CGI
CpG island
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher's web-site
References
- 1.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 2.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 4.Kelso A, Gough NM. Coexpression of granulocyte-macrophage colony-stimulating factor, gamma interferon, and interleukins 3 and 4 is random in murine alloreactive T-lymphocyte clones. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9189–9193. doi: 10.1073/pnas.85.23.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paliard X, de Waal Malefijt R, Yssel H, Blanchard D, Chretien I, Abrams J, de Vries J, et al. Simultaneous production of IL-2, IL-4, and IFN-gamma by activated human CD4 +and CD8+ T cell clones. J. Immunol. 1988;141:849–855. [PubMed] [Google Scholar]
- 6.Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Panzer M, Sitte S, Wirth S, Drexler I, Sparwasser T, Voehringer D. Rapid in vivo conversion of effector T cells into Th2 cells during helminth infection. Journal of Immunology. 2012;188:615–623. doi: 10.4049/jimmunol.1101164. [DOI] [PubMed] [Google Scholar]
- 8.Peine M, Rausch S, Helmstetter C, Frohlich A, Hegazy AN, Kuhl AA, Grevelding CG, et al. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, Can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 2013;11:e1001633. doi: 10.1371/journal.pbio.1001633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohning M, Grogan JL, Coyle AJ, Yazdanbakhsh M, Meisel C, Gutierrez-Ramos JC, Radbruch A, et al. T1/ST2 expression is enhanced on CD4+ T cells from schistosome egg-induced granulomas: analysis of Th cell cytokine coexpression ex vivo. J. Immunol. 1999;162:3882–3889. [PubMed] [Google Scholar]
- 10.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat. Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee D, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 12.Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc. Natl. Acad. Sci. U.S.A. 1982;79:3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr ARW, James KD, Turner DJ, Smith C, et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce EJ, MacDonald AS. The immunobiology of schistosomiasis. Nat. Rev. Immunol. 2002;2:499–511. doi: 10.1038/nri843. [DOI] [PubMed] [Google Scholar]
- 18.Henderson GS, Lu X, McCurley TL, Colley DG. In vivo molecular analysis of lymphokines involved in the murine immune response during Schistosoma mansoni infection. II. Quantification of IL-4 mRNA, IFN-gamma mRNA, and IL-2 mRNA levels in the granulomatous livers, mesenteric lymph nodes, and spleens during the course of modulation. J. Immunol. 1992;148:2261–2269. [PubMed] [Google Scholar]
- 19.Wynn TA, Eltoum I, Cheever AW, Lewis FA, Gause WC, Sher A. Analysis of cytokine mRNA expression during primary granuloma formation induced by eggs of Schistosoma mansoni. J. Immunol. 1993;151:1430–1440. [PubMed] [Google Scholar]
- 20.Brunet LR, Finkelman FD, Cheever AW, Kopf MA, Pearce EJ. IL-4 protects against TNF-alpha-mediated cachexia and death during acute schistosomiasis. J. Immunol. 1997;159:777–785. [PubMed] [Google Scholar]
- 21.Fallon PG, Richardson EJ, McKenzie GJ, McKenzie AN. Schistosome infection of transgenic mice defines distinct and contrasting pathogenic roles for IL-4 and IL-13: IL-13 is a profibrotic agent. J. Immunol. 2000;164:2585–2591. doi: 10.4049/jimmunol.164.5.2585. [DOI] [PubMed] [Google Scholar]
- 22.Booth M, Mwatha JK, Joseph S, Jones FM, Kadzo H, Ireri E, Kazibwe F, et al. Periportal fibrosis in human Schistosoma mansoni infection is associated with low IL-10, low IFN-gamma, high TNF-alpha, or low RANTES, depending on age and gender. J. Immunol. 2004;172:1295–1303. doi: 10.4049/jimmunol.172.2.1295. [DOI] [PubMed] [Google Scholar]
- 23.Blanton RE, Salam EA, Ehsan A, King CH, Goddard KA. Schistosomal hepatic fibrosis and the interferon gamma receptor: a linkage analysis using single-nucleotide polymorphic markers. Eur. J. Hum. Genet. 2005;13:660–668. doi: 10.1038/sj.ejhg.5201388. [DOI] [PubMed] [Google Scholar]
- 24.Dessein AJ, Hillaire D, Elwali NE, Marquet S, Mohamed-Ali Q, Mirghani A, Henri S, et al. Severe hepatic fibrosis in Schistosoma mansoni infection is controlled by a major locus that is closely linked to the interferon-gamma receptor gene. Am. J. Hum. Genet. 1999;65:709–721. doi: 10.1086/302526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor JJ, Krawczyk CM, Mohrs M, Pearce EJ. Th2 cell hyporesponsiveness during chronic murine schistosomiasis is cell intrinsic and linked to GRAIL expression. J Clin Invest. 2009;119:1019–1028. doi: 10.1172/JCI36534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 28.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 29.Phythian-Adams AT, Cook PC, Lundie RJ, Jones LH, Smith KA, Barr TA, Hochweller K, et al. CD11c depletion severely disrupts Th2 induction and development in vivo. J. Exp. Med. 2010;207:2089–2096. doi: 10.1084/jem.20100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.