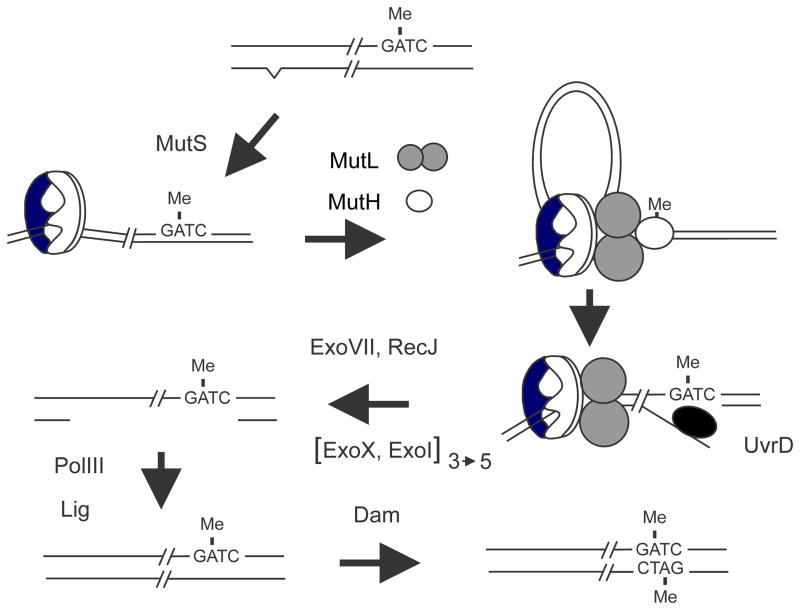
FIGURE 6.
Dam-directed mismatch repair in E. coli. The top of the figure shows DNA immediately behind the replication fork in which the “old” top strand is methylated and the “new” strand is not and also contains a base mismatch (carat) created as a replication error. The mismatch is recognized and bound by MutS followed by recruitment of MutL and MutH to form a ternary complex. The formation of this complex is thought to involve DNA looping to bring the mismatch and a GATC sequence in close proximity but the details are unclear. In the ternary complex the latent nuclease activity of MutH is activated and it cleaves the new unmethylated strand 5′ to the GATC sequence. The nick created by MutH serves as an entry site for the UvrD helicase which unwinds the DNA exposing single-stranded DNA which is digested by one or more of the following exonucleases: RecJ, ExoVII, ExoX or ExoI. The exonuclease(s) used depends on the relative orientation of the mismatch to the GATC sequence; in the figure the direction of UvrD unwinding is 5′ to 3′ and so either ExoVII or RecJ or both are needed. If the mismatch was to the “right” of the GATC sequence, UvrD would unwind in the 3′ to 5′ direction and ExoX and/or ExoI would digest the single-stranded DNA. The gap created by nuclease digestion removes the mismatched base and is filled in by DNA polymerase III. The resulting nick is closed by DNA ligase and eventual Dam methylation precludes any further repair.