Fig. 6.
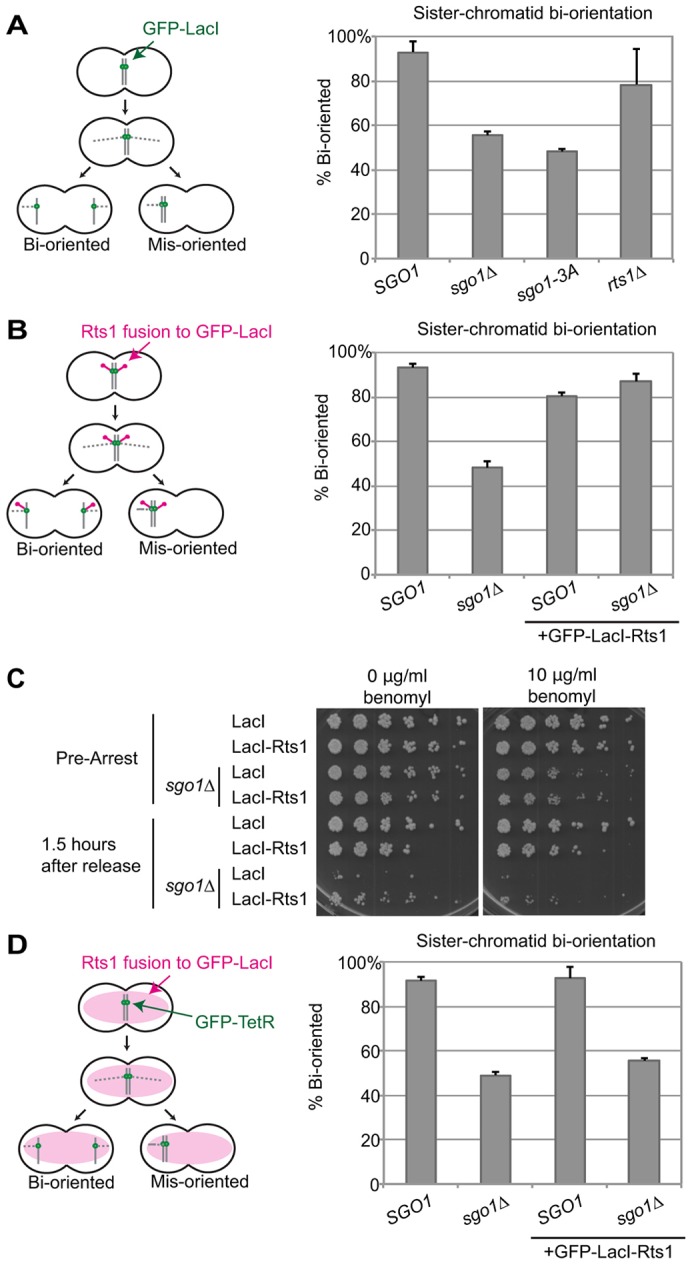
Recruitment of Rts1 is sufficient for bi-orientation. Bi-orientation of sister chromatids after release from benomyl. (A) sgo1Δ or sgo1-3A mutants fail to properly bi-orient after release from benomyl. Cells containing a GFP–LacI/LacO dot near the centromere of chromosome V were arrested in medium containing benomyl to promote spindle disassembly. Cells were then released into medium without benomyl to allow the spindle to reform. After 60 min, cells were fixed and stained with DAPI to visualize DNA. Only cells that had fully segregated their DNA were scored for localization of chromosome V GFP dots (n≥48 cells). Results are the mean of two experiments, with error bars representing the standard deviation. (B) A GFP–LacI–Rts1 fusion protein rescues the bi-orientation defect of sgo1Δ cells. The same bi-orientation assay as in A was performed using a GFP–LacI–Rts1 fusion in place of GFP–LacI. (C) GFP–LacI–Rts1 does not restore normal proliferation to cells lacking SGO1. Cells were arrested in benomyl and then released into medium without benomyl. After 90 min, cells were plated in serial dilutions on media with or without benomyl as indicated. (D) The GFP–LacI–Rts1 fusion protein restores bi-orientation to sgo1Δ cells only when localized to the centromere. A GFP–LacI–Rts1 fusion protein was expressed as in B, but in this case there were no LacO arrays on the chromosome. Chromosome segregation was instead monitored with a TetR–GFP/TetO dot on chromosome V.
