Figure 11. Structural/modeling studies of TLS polymerases bound to processivity clamps.
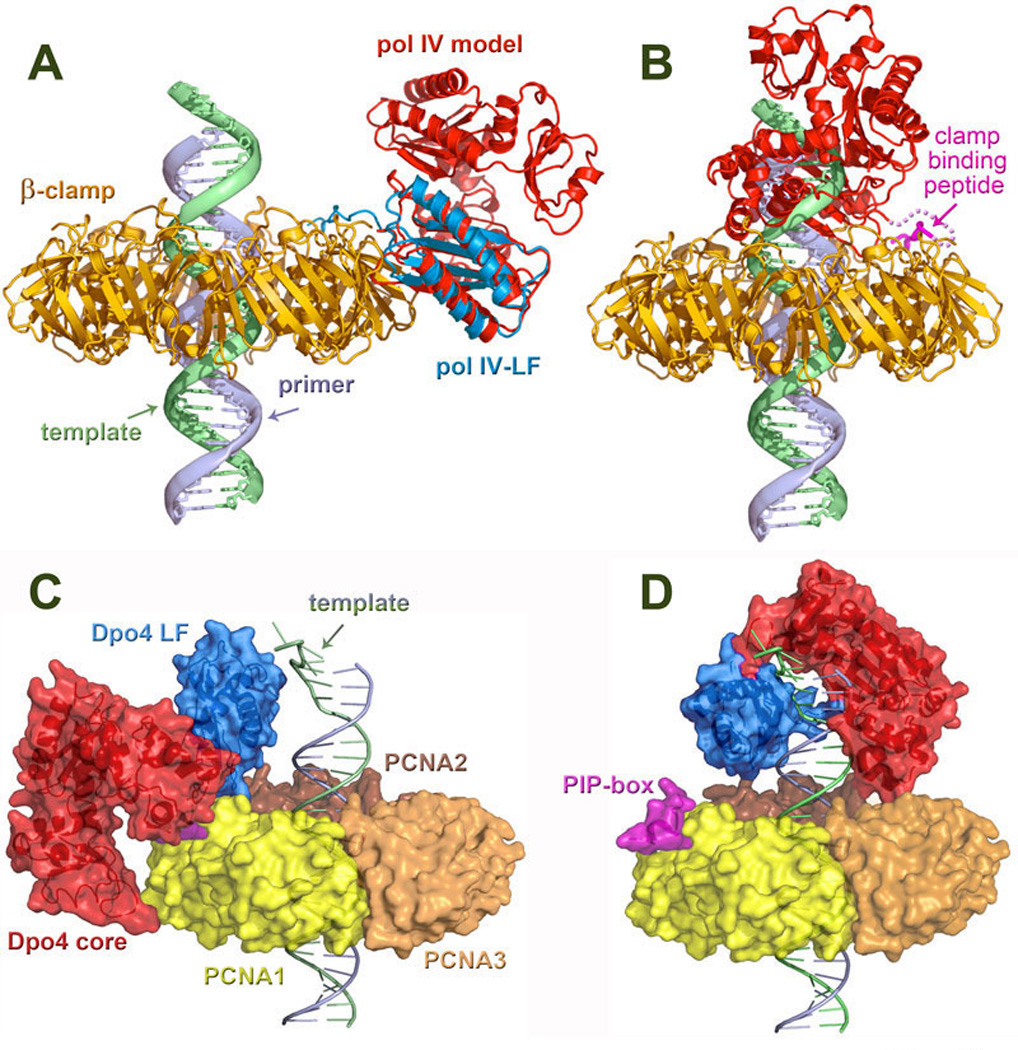
A. Model of a pol IV (shown in red) bound to the β-clamp (gold) in an inactive position. The position of the polymerase was modeled by superimposing the little finger domain from the ternary complex of Dpo4 with DNA and an incoming nucleotide (PDB code: 1JXL) (133) onto the LF of E. coli pol IV (blue) in a complex with the β-clamp (PDB: 1UNN) (38). In this position, pol IV is unable to access the primer/template terminus. B. Model of pol IV bound to the β-clamp and DNA at the primer/template junction following a polymerase switch. The position of the polymerase was modeled by superimposing the DNA from the ternary complex of Dpo4 onto the end of a DNA molecule running perpendicularly through the β-clamp. Contact with the clamp is maintained by the C-terminal clamp-binding peptide (pink), which tethers the polymerase to the replication complex. C. Dpo4 in an inactive extended form bound to PCNA (PDB: 2NTI) (261) with dsDNA passing through its central aperture. In this conformation, the core (red) and little finger (blue) of Dpo4 contact PCNA (the trimer is shown in yellow, orange, and brown) to facilitate the PIP (pink)–PCNA1 (yellow) binding. D. Model of Dpo4 in an active form bound to a DNA template and a PCNA ring. Similar to the structure shown in the panel C, in this conformation the PIP-box anchors Dpo4 to PCNA1. The DNA-bound Dpo4 is modeled onto PCNA from the type I structure (PDB: 1JX4). Panels A and B reproduced with permission from Bunting et al., (38). Panels C and D were provided through the courtesy of Dr. Hong Ling, University of Western Ontario, London, Canada. All four structures provide structural support for the proposed “tool-belt” model for polymerase switching (176).
