Figure 12. Comparison of the crystal and NMR structure of UmuD'2.
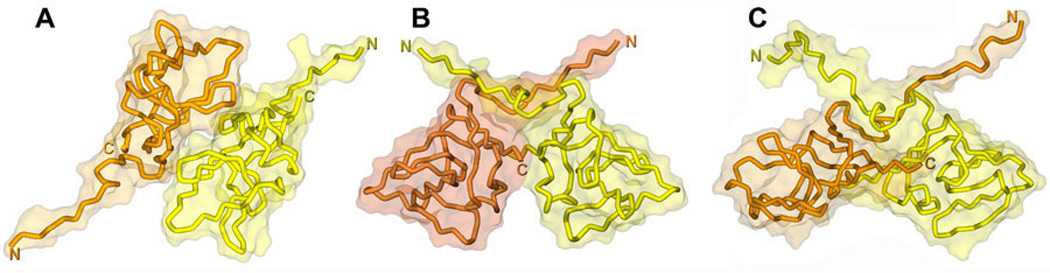
A. Crystal structure of a UmuD' “Molecular” dimer (PDB: 1AY9) in which the N-termini of each protomer are extended in diametrically opposite directions (182). B. Crystal structure of UmuD' “Filament” dimer (PDB: 1UMU) in which the N-terminal tail of each protomer cross over the globular body of its partner in a configuration that is likely to mimic the one in which the RecA-mediated cleavage of UmuD to UmuD' occurs (183). C. NMR solution structure of UmuD'2 (PDB: 1I4V) revealing a structural fold similar to the crystallographic “Filament” dimer, but in which the globular domain is somewhat “flattened” and the N-terminal tails are free in solution (64). The protein backbone is depicted as a tube that is surrounded by the semi-transparent solvent accessible surface of the dimer. One UmuD' protomer is colored yellow, while the other one is in orange. The structures were created using Discovery Studio Visualizer.
