TABLE 1.
Summary of expression and biochemical data on PGM1 deficiency mutants
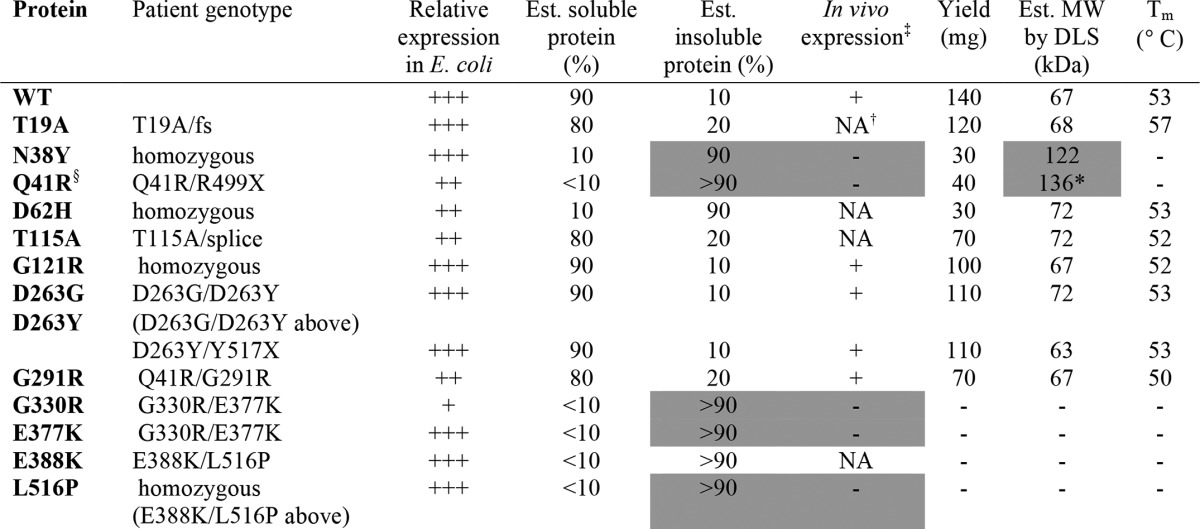
‡ In vivo data on protein expression levels obtained from patient fibroblasts as described in Ref. 4; NA indicates sample not available.
† Description of in vivo protein expression protein for T19A in Ref. 3 is ambiguous, so we list as NA. Alleles with frame shift (fs) or premature stop codons (X) are unlikely to contribute to in vivo protein expression levels; for detailed information on patient genotypes, see Ref. 3.
§ Additional genotype for this mutant's shown under G291R; good in vivo protein expression in sample heterozygous for Q41R/G291R is consistent with reduced expression of Q41R but normal expression of the G291R variant.
* Data shown for this sample is post-purification by size exclusion chromatography. Relative expression in E. coli reflects a combination of soluble and insoluble protein fractions. Gray shading highlights the correlation between poor in vivo expression of certain PGM1 mutants with poor solubility/aggregation of recombinantly expressed proteins. The G291R mutant was also characterized in a patient heterozygous for a fs mutant (1), where it showed reduced but visible protein expression in vivo.
