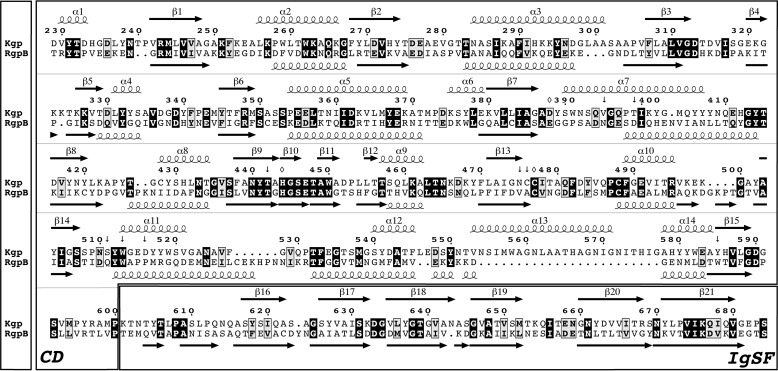
FIGURE 1.
Gingipain sequences. Structure-guided sequence alignment of the CD and IgSF moieties (separately framed) of Kgp from P. gingivalis strain W83 (UniProt accession number Q51817; top rows) and RgpB from P. gingivalis strain HG66 (GenBank accession number AAB41892; Protein Data Bank code 1CVR (28); bottom rows) is shown. The sequence of the latter differs from that of the ortholog from strain W83 at 12 positions (UniProt accession number 95493; Protein Data Bank code 4IEF (27)). The amino acid numbering and the regular secondary structure elements (strands as black arrows labeled β1–β21 and helices as loops labeled α1–α14) above the alignment correspond to the Kgp(CD+IgSF) structure (this study); those below the alignment correspond to RgpB(CD+IgSF) (taken from Fig. 2c of Ref. 28). Identical residues are in bold white over black background, similar residues are in bold black over gray background, and the overall sequence identity is 27%. (Potential) catalytic residues of Kgp CD are pinpointed by an open rhombus; residues framing the S1 pocket are indicated by an arrow.