Figure 1.
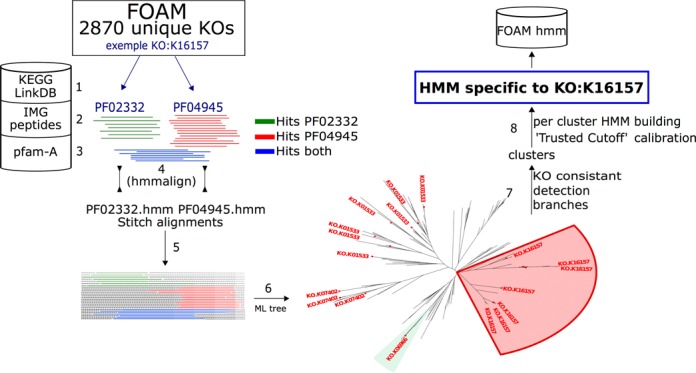
HMM building pipeline: example with KO:K16157 (methane monooxygenase). Step 1—find Pfam(s) combination assigned to the KO of interest (a) and (b) check for redundancy. Step 2—fetch IMG peptide sequences which hit the retrieved Pfam(s). Step 3—fetch from Pfam-A database the HMM of interest. Step 4—alignment (hmmalign) and filter each Pfam from extra sequences obtained in IMG. Step 5—stitch filtered alignments. Step 6—draw a Maximum Likelihood tree (fasttree). Step 7—find clusters in tree with same KO. Step 8—split alignment (step 5 output) by cluster (step 7 output) and build HMM for each, and process the ‘Trusted Cutoff’ computation.
