Figure 8.
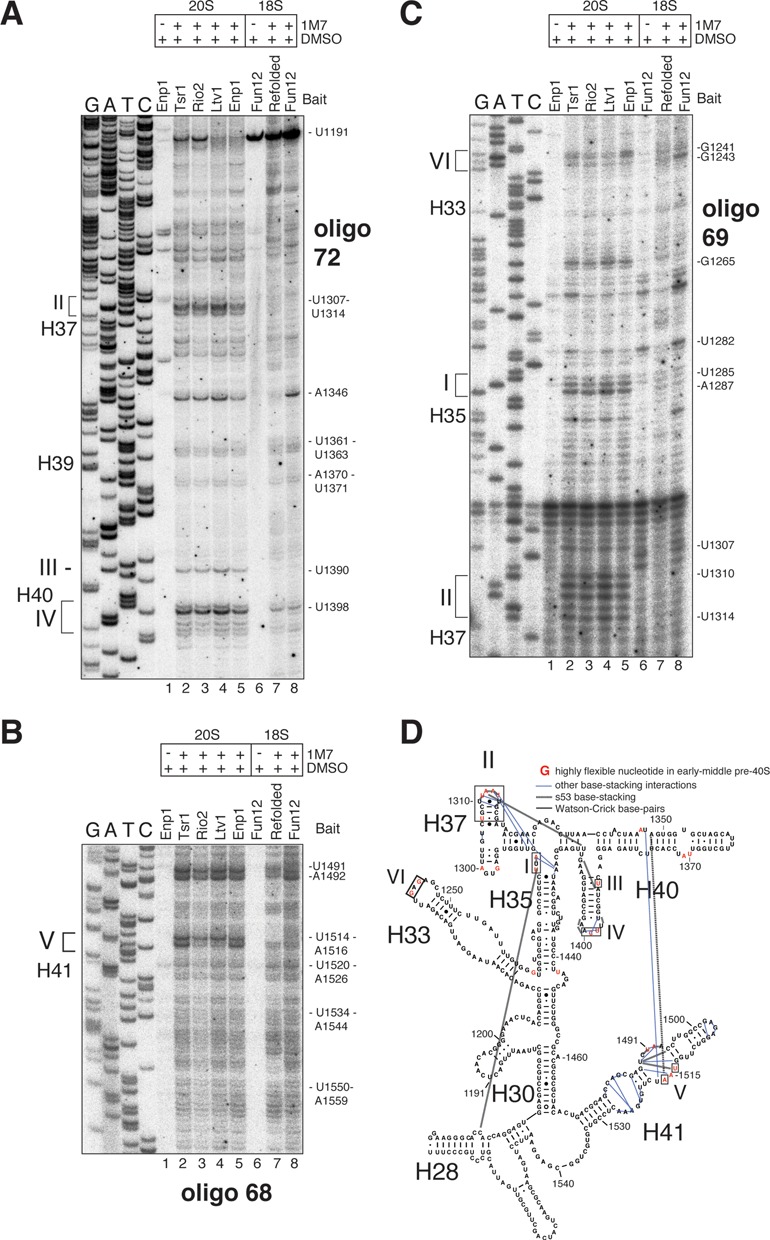
Ribosome assembly factors are required to maintain an open head domain conformation. (A–C) Comparison of primer extension data obtained from 1M7 modified early (Ltv1, Enp1), middle (Tsr1, Rio2) pre-40S complexes (lanes 2–5), mature 40S subunits (Fun12) (lane 8) and in vitro refolded 18S rRNA (lane 7, Refolded). Unmodified 20S and 18S rRNAs were used as control samples (lanes 1 and 6). The positions of the modified nucleotides are indicated on the right side of each panel. (D) Overview of TCPEM output generated from in vitro refolded 18S rRNA ChemModSeq data. Shown are the results for the 3′ major domain. Yellow letters indicate nucleotides that the algorithm predicted were most likely 1M7 modified.
