Figure 7.
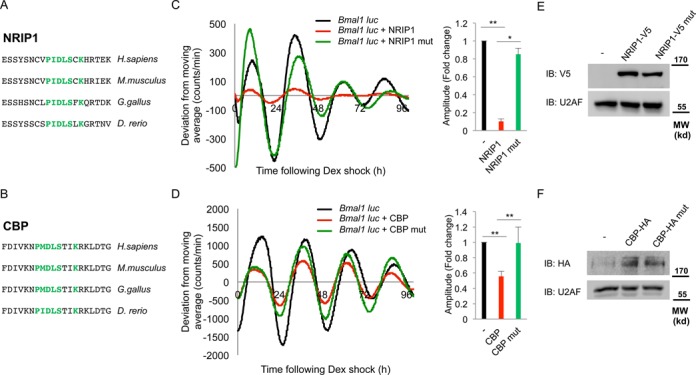
The PXDLS-containing proteins, NRIP1 and CBP, affect circadian rhythmicity in a PXDLS-dependent manner. Schematic representation of the evolutionary conservation of the PXDLS motif (green) in (A) NRIP1 and (B) CBP. NIH3T3 cells were transiently transfected with Bmal1-luciferase reporter either with control empty vector (black) or with (C) wild-type NRIP1-V5 (red) or PXDLS mutant NRIP1-V5 (green), (D) wild-type CBP-HA (Red) or PXDLS mutant CBP-HA (green). Cells were synchronized with a short dexamethasone (Dex) treatment, and real-time bioluminescence recording was performed using the LumiCycle. Amplitudes were quantified using the LumiCycle software. Data are presented on a bar graph as fold change (mean +/− SD, n = 4), P-values < 0.001 is marked with ** and <0.005 is marked with *. The expression levels of (E) wild-type NRIP1-V5 and PXDLS mutant NRIP1-V5 and (F) wild-type CBP-HA and PXDLS mutant CBP-HA were determined by SDS-PAGE and IB. U2AF was used as loading control. Molecular Weight (MW).
