Figure 4.
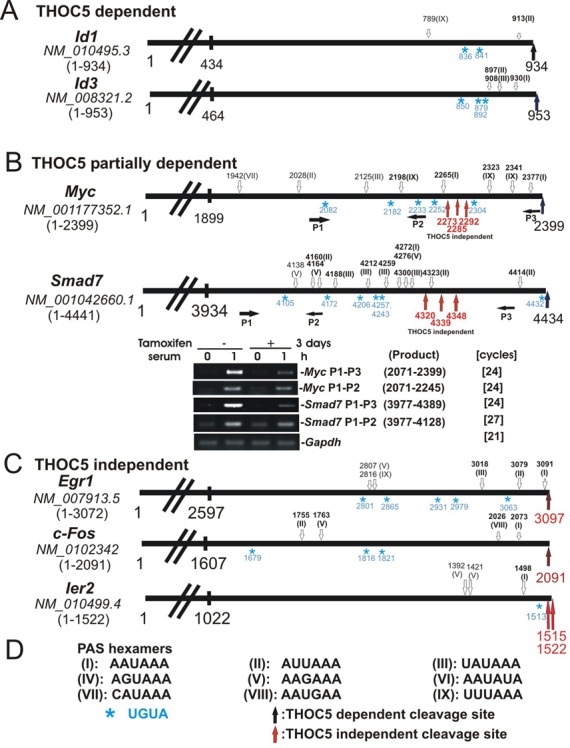
In the absence of THOC5, processing of Myc or Smad7 genes was partially rescued by a backup pathway but cleaved ∼100 nt upstream from the cleavage site in the presence of THOC5. Schematic presentation of 3′UTR of several IEGs and their accession numbers (italic). Numbers represent nucleotide numbers without poly A. . PAS hexamers (white arrows; numbers represent the first nucleotide of PAS sequence shown in (D). Common PAS hexamers in mouse and human were shown in bold and the UGUA element was shown ‘blue *’. THOC5 dependent cleavage sites are presented by black arrows; THOC5 independent cleavage sites are represented by red arrows. (A) 3′UTR of THOC5 dependent genes, Id1 and Id3. (B) 3′UTR of Myc and Smad 7 genes: both genes contain THOC5 independent cleavage sites. Semi-quantitative Myc and Smad7-specific RT-PCR using up- (P1–P2) or down- stream (P1–P3) of THOC5 independent cleavage sites in the presence or absence of THOC5. (C) THOC5 independent gene (the most rapidly induced IEG). (D) Sequences of PAS hexamers that were indicated (21) in (A–C) as I-IX.
