Figure 3.
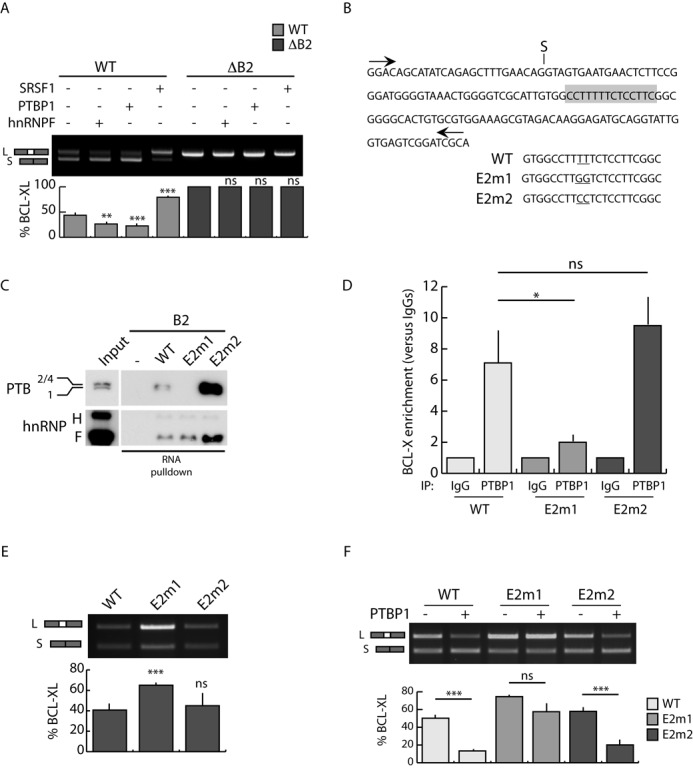
The B2 element is required for PTBP1-dependent BCL-Xs splicing. (A) RT-PCR analysis of the in vivo splicing assays performed in HEK293T cells transfected with WT or mutated (ΔB2) BCL-X minigenes and the indicated splicing factors. The bar graph of the percentage of BCL-XL is also shown (mean ± SD, n = 3). (B) Scheme of BCL-X exon 2 sequence. The putative PTBP1 binding site is highlighted (gray box). The mutated bases in E2m1 and E2m2 mutants are underlined. (C) Western blot analysis of RNA-pulldown assay using biotin-labeled BCL-X WT or mutant (E2m1 and E2m2) RNA. Streptavidin beads have been used as control (−). (D) CLIP experiments of PTBP1 performed in HEK293T transfected with the indicated minigenes. Associated BCL-X RNA was quantified by qPCR (primers used are indicated in Supplementary Table S1). Data are represented as fold enrichment relative to the IgG sample (mean ± SD, n = 3). (E and F) RT-PCR analysis of in vivo splicing assays performed in HEK293T cells transfected with wt and mutated (E2m1 and E2m2) minigenes in the presence (F) or absence (E) of PTBP1. The bar graph of the percentage of BCL-XL is shown (mean ± SD, n = 3). (A, D, E and F) The P values of Student's t-test are reported. *P < 0.05, **, P < 0.01; ***, P < 0.001; n.s., not significant.
