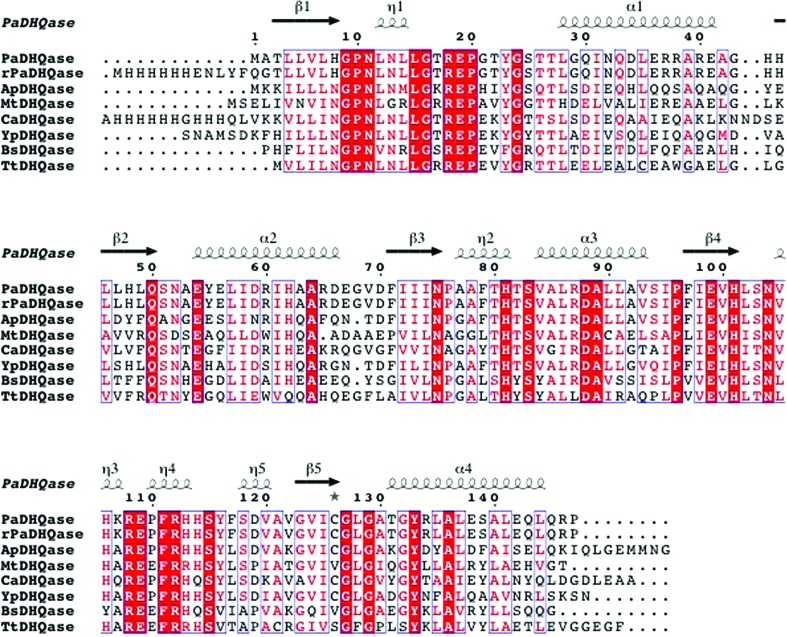
Figure 4.
Comparison of PaDHQase with other type II DHQases. The amino-acid sequence alignment showing structural elements of DHQase was generated with ESPript3.0 (Gouet et al., 2003 ▶). Secondary-structure elements are as follows: α-helices are shown as large squiggles labeled α, 310-helices are shown as small squiggles labeled η, β-strands are shown as arrows and are labeled β. Identical residues are shown on a red background, conserved residues are shown in red and conserved regions are shown in blue boxes. The DHQases used in alignment were PaDHQase, recombinant PaDHQase (rPaDHQase), and others identified using the Structure Similarity option of PDBeFold (http://www.ebi.ac.uk/msd-srv/ssm/) as most structurally similar to PaDHQase. ApHHQase is the crystal structure of DHQase from Actinobacillus pleuropneumoniae (PDB entry 1uqr), the most similar to PaDHQase, and has 66% sequence identity and an r.m.s.d. 0.63 Å (Maes et al., 2004 ▶). YpDHQase, the crystal structure of the homolog from Yersinia pestis (PDB entry 3lwz), is the next closest structural homolog with 68% sequence identity and an r.m.s.d. of 0.75 Å (Center for Structural Genomics of Infectious Diseases, unpublished work). CaDHQase is the homolog from Candida albicans (PDB entry 3kip), which has 56% sequence identity and an r.m.s.d. of 0.76 Å (Trapani et al., 2010 ▶). Other structurally similar type II DHQases include MtDHQase from M. tuberculosis (Gourley et al., 1999 ▶; Schmidt et al., 2013 ▶), BaDHQase from Bacillus subtilis (PDB entry 1gqo; D. A. Robinson, A. W. Roszak, J. R. Coggins & A. J. Lapthorn, unpublished work), TtDHQase from Thermus thermophilus (PDB entry 2uyg; H. Utsunomiya, Y. Agari, T. Imagawa & H. Tsuge, unpublished work) and ScDHQase from S. coelicolor (Roszak et al., 2002 ▶).