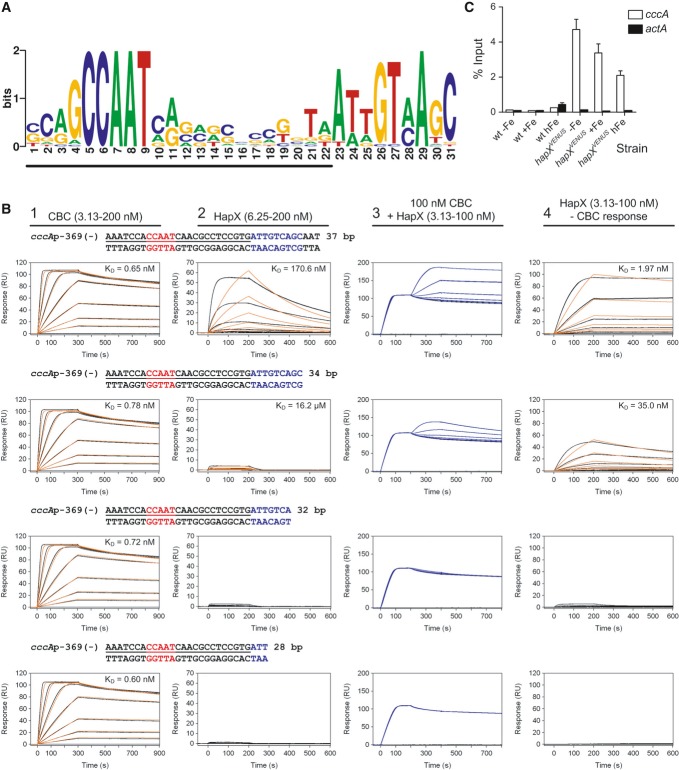
Figure 6. HapX binds in vitro and in vivo to an evolutionary conserved motif identified in promoters of cccA homologs.
- An evolutionary conserved, bipartite motif in promoters of cccA homologs identified by MEME analysis. The underlined nucleotides would be covered upon CBC-binding as can be predicted based on the identified CBC/DNA binary complex crystal structure (Huber et al, 2012).
- Real-time SPR characterization of in vitro formation of the CBC/DNA-HapX ternary complex on the conserved cccA promoter motif from A. fumigatus. SPR analyses included binding of the CBC to DNA (panel 1), HapX to DNA (panel 2) and HapX to preformed CBC/DNA complexes (panel 3). The SPR sensorgrams are shown from sensor-immobilized 37 base pair duplexes covering the full as well as 3′-truncated duplexes. Nucleotides marked in blue represent the HapX consensus binding site in fungal cccA promoters identified by MEME analysis. Binding responses of the indicated CBC or HapX concentrations injected in duplicate (black lines) are shown overlaid with the best fit derived from a 1:1 interaction model including a mass transport term (red lines). Binding responses of CBC/DNA-HapX ternary complex formation (panel 3, blue lines) were obtained by concentration-dependent co-injection of HapX on preformed binary CBC/DNA complexes after 200 s within the steady-state phase. Sensorgrams in panel 4 depict the association/dissociation responses of HapX on preformed CBC/DNA and were generated by CBC response (co-injection of buffer instead of HapX) subtraction from HapX co-injection responses. Dissociation constants (KD) are plotted inside the graphs.
- ChIP analysis demonstrating in vivo binding of HapX to the conserved cccA promoter motif from A. fumigatus. ChIP qPCR was performed on wild-type or the strain containing Venus-tagged HapX (hapXVENUS) grown for 18 h, then shifted to fresh media with no iron (−Fe), 0.03 mM iron (+Fe), or 3 mM iron (hFe) for 8 h. DNA was immunoprecipitated with either a control IgG antibody, or anti-GFP polyclonal antibody that recognizes the Venus protein. Binding of HapXVENUS to the DNA region was assessed by qPCR. HapX binding is represented as percent enrichment of input control samples ± SD from triplicates. The actA (actin) promoter served as a negative control.