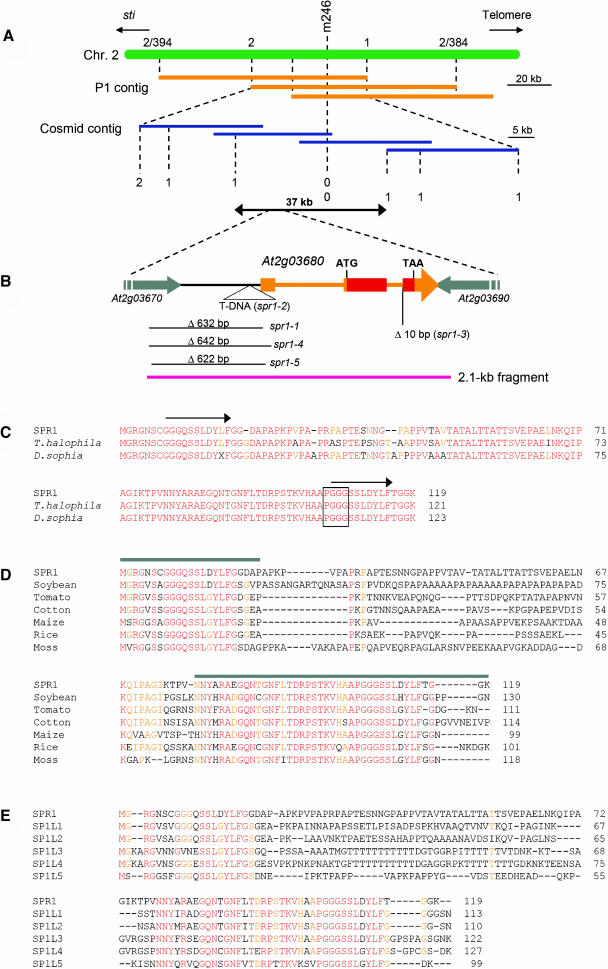
Figure 2.
Cloning Procedure and Protein Sequence Alignments of SPR1.
(A) Fine mapping of the SPR1 locus. The region flanking the CAPS marker m246 was covered by P1 phage clones (orange bars). CAPS markers were created from the P1 sequences and used to locate the SPR1 locus to a 60-kb region. This region was covered by cosmid clones (blue bars), and new CAPS markers were generated from the cosmid sequences. Mapping with these markers finally confined the SPR1 locus to a 37-kb region. Numbers of recombination events are shown.
(B) Genomic region surrounding the SPR1 gene (At2g03680). Transcribed region of SPR1 is colored orange or red. Orange boxes represent exons, and thin orange bars represent introns. The SPR1-coding region is red. Parts of the two flanking genes are shown with gray arrows. Molecular lesions found in five spr1 mutant alleles are shown. The pink bar represents the 2.1-kb genomic fragment shown to complement spr1-1 mutants.
(C) Protein sequence alignment of SPR1 and two Brassicaceae family homologs. Residues identical in all three proteins are shown in red, and those identical in two proteins are shown in orange. Arrows above the SPR1 sequence indicate peptide segments with sequence identity. The PGGG motif found in some mammalian MAPs is boxed.
(D) Protein sequence alignment of SPR1 and six non-Brassicaceae family homologs. Residues identical in all seven proteins are shown in red, and those identical in five or six proteins are orange. Gray bars above the SPR1 sequence indicate the region with high sequence identity among all SPR1 homologs. These regions were used for the GFP fusion experiments shown in Figure 5.
(E) Protein sequence alignment of SPR1 and five Arabidopsis homologs. Residues identical in all six proteins are shown in red, and those identical in five are orange.
Accession numbers for the sequences used in the alignments are listed in Methods.