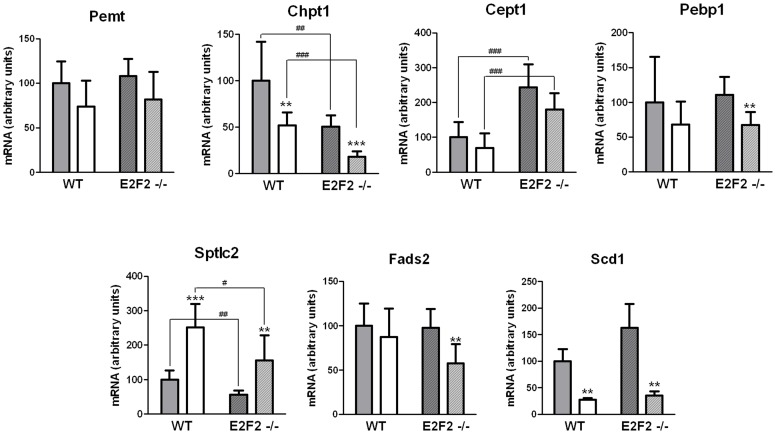
Figure 4. E2F2 gene deletion promotes changes in transcript expression of selected genes involved in phospholipid and fatty acid metabolism.
Partial hepatectomy was performed on E2F2+/+ (wild-type, WT) and E2F2-/- mice, and were sacrificed 48 hours later. Quiescent (0-h, solid bars) and regenerating (48-h, open bars) livers were harvested and total RNA from each mouse was examined in triplicate by quantitative real-time PCR for the following genes: Pemt, phosphatidylethanolamine N-methyltransferase; Chpt1, choline phosphotransferase 1; Cept1, choline/ethanolamine phosphotransferase 1; Pebp1, phosphatidylethanolamine binding protein 1; Sptlc2, serine palmitoyltransferase, long chain base subunit 2; Fads2, fatty acid desaturase 2; Scd1, stearoyl-CoA desaturase 1. Relative transcript abundance was calculated for each mouse using a normalization factor computed by Genorm software for pyruvate carboxylase, transferrin receptor and vascular endothelial zinc finger containing factor 1 mRNAs. Values are referred to 0-h WT mice samples (100%). Results are presented as means ± SD (n = 8 mice per group), except for Scd1 transcript levels, which were estimated in 3 pools of eight mice each from each genotype (n = 3). Statistical differences between regenerating and quiescent liver of a genotype are denoted by ** P≤0.01, *** P≤0.001, and between the same tissue condition of different genotypes are denoted by # P≤0.05, ## P≤0.01, ### P≤0.001 (unpaired, 2-tailed Student's t-test).