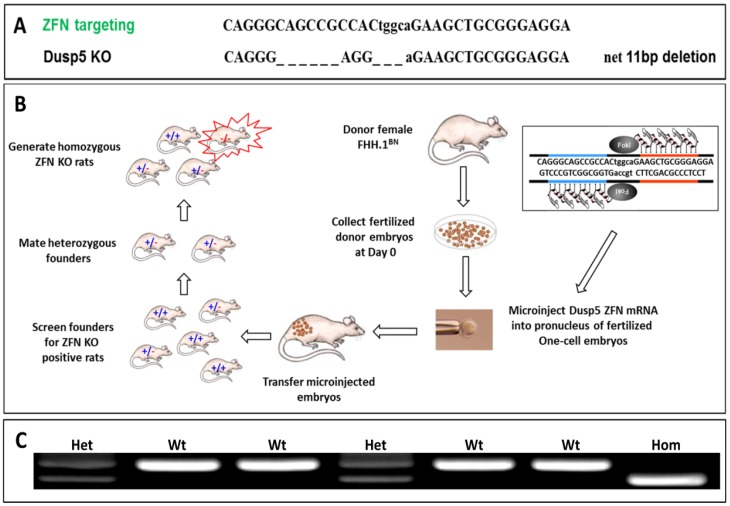
Figure 4. Schematic describing the generation of Dusp5 ZFN KO rats in the FHH.1BN genetic background.
Panel A presents the sequence of the Dusp5 ZFN. The Dusp5 specific ZFN introduced a 14 bp deletion and a 3 bp insertion resulting in a net 11 bp deletion between nucleotides 449–464 in the Dusp5 sequence in the KO animals that introduced a frame shift mutation. Panel B presents the strategy for the generation of Dusp5 ZFN KO rats in the FHH.1BN genetic background. Fertilized donor embryos from female FHH.1BN rats were collected and the Dusp5 ZFN mRNA was microinjected into pronuclei of the fertilized one-cell embryos. These embryos were transferred back to a foster mother. The heterozygous founders were brother-sister mated to generate the homozygous ZFN KO founders and the FHH.1BN wild type control rats. Panel C presents an example of PCR genotyping of the region of interest in FHH.1BN and Dusp5 KO rats. The rats were genotyped using the following primers: Dusp5-F: 5′-GCT GCA GGA GGG CGG CGG CG -3′, Dusp5 R: 5′-CTT TAA GGA AGT AGA CCC G-3′. These primers amplify a 155 bp band for the wild type allele and a 144 bp band for the knockout allele. Both bands are observed in heterozygous rats.