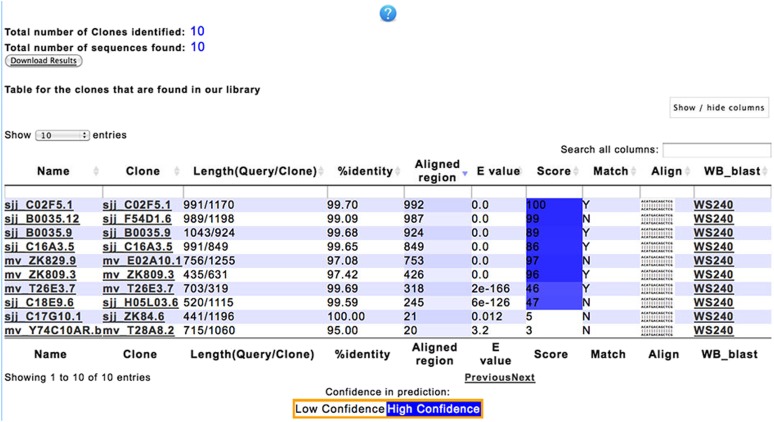
Figure 3.
An example of RNAi clone identification using Clone Mapper. The DNA sequences obtained upon sequencing of 10 RNAi clones, from (Zugasti et al. 2014), were used as input into Clone Mapper. The results obtained, ranked by “Aligned region,” are shown in this screen-grab. The leftmost column shows the library name of each clone, the next column the name of the clone that best matches the experimentally determined RNAi clone insert sequence. In this example, half the clones appeared to be what was expected; for 3 of 5 of the others, an alternative identity was assigned with high confidence. For the remaining clones only a very short sequence matches a clone in the in silico library. These sequences can be compared directly to the genome of C. elegans by clicking the link in the rightmost column. The exact meaning of the different columns and options is explained in the help document, accessible by clicking the question mark at the top of the screen.