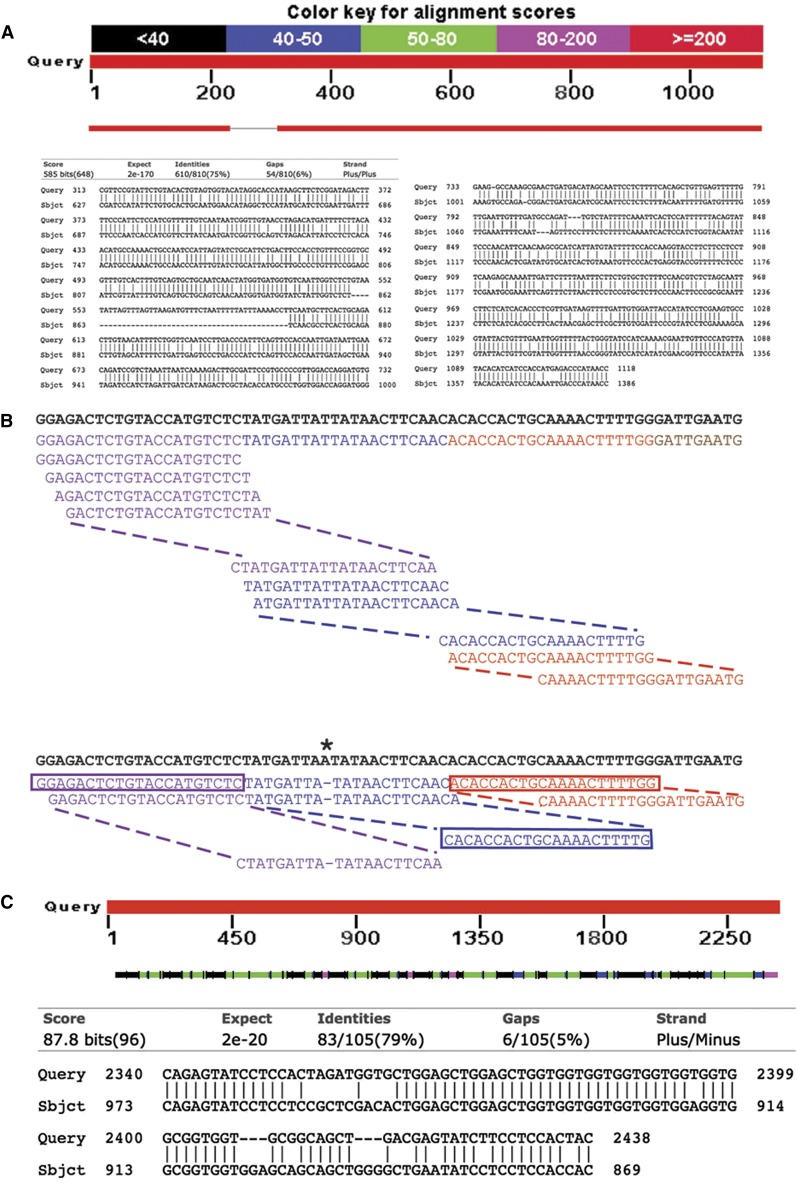
Figure 4.
Basis of the target identification strategy. (A) An example of a target identified by Wormbase but not Clone Mapper. The RNAi clone-transcript pair (sjj_B0222.9 - F15E6.6) displays overall high identity, with blocks of >200 nucleotides with >80% identity, but does not contain a single 21 bp contiguous stretch of identical sequence. (B) The approach implemented in Clone Mapper for defining targets of RNAi clones. The set of possible nonoverlapping 21mer fragments (PNO) are generated starting from the 5′ end of each predicted clone insert sequence (in black). In the example shown, there are 3 complete (purple, blue, red) and one partial (brown) PNOs. All possible overlapping 21mer segments (POS) are generated and assigned to the corresponding PNO; for simplicity only a selection of POS are shown for each PNO. The library of all transcripts is queried with each POS to identify matched overlapping segments (MOS). In the example shown in the lower part of the panel, a transcript (in bold) with a single difference from the clone insert sequence (*) is shown. The number of PNOs that contain at least one POS that exactly matches a given transcript is counted (MNO; here 3). An example of one matching POS for each PNO is boxed. A score is then calculated (see Materials and Methods). (C) An example of a target identified by Clone Mapper but not Wormbase. Here the RNAi clone insert sequence has multiple stretch of sequence that have perfect matches over more than 21 nt to a target transcript (upper part of the panel; sjj_Y50E8.g and ZK643.8a), but no contiguous region of 100 nt with 95% identity (lower part of the panel; the longest stretch of identity in the selected alignment of one fragment shown here is 31 nt).