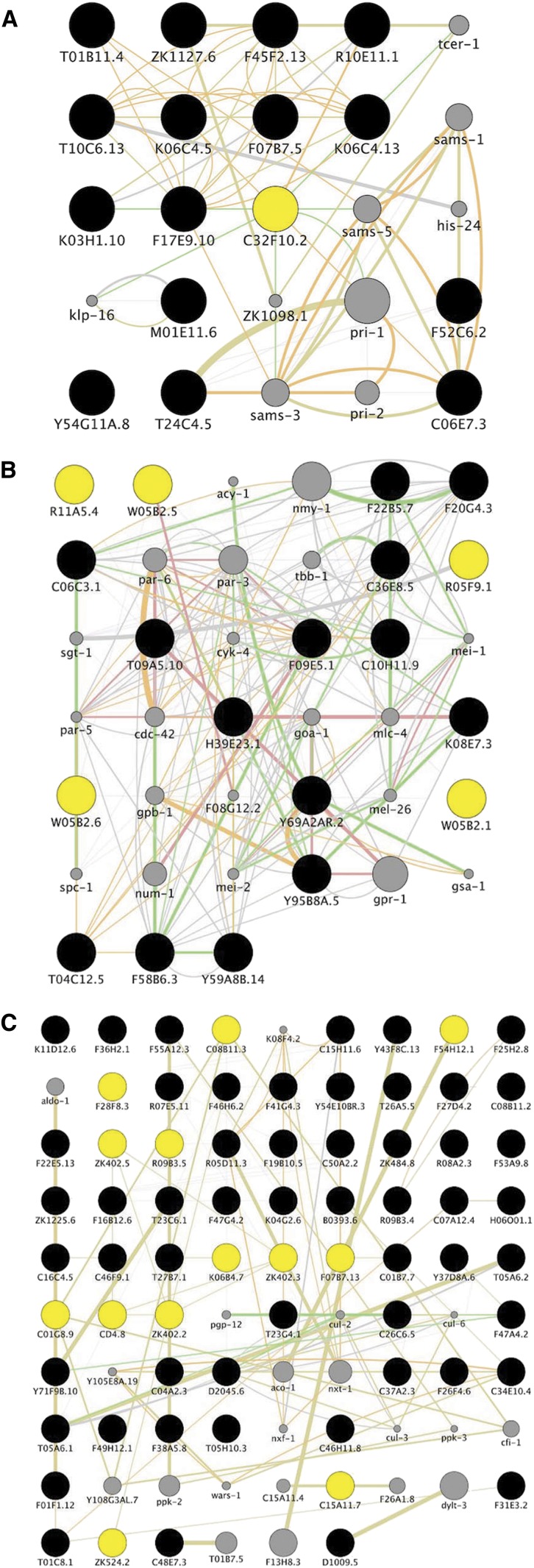
Figure 6.
Network analysis of novel RNAi targets. (A) Ceron et al. undertook an RNAi screen to identify genes that interact with the C. elegans retinoblastoma gene lin-35 (Ceron et al. 2007). The list of novel targets identified with Clone Mapper for the RNAi clones selected by Ceron et al. was used as input to GeneMania (black circles), together with lin-35/C32F10.2 (highlighted in yellow) as a seed gene. (B) Fievet et al. performed RNAi screens for C. elegans cell polarity mutants, to generate a polarity network (Fievet et al. 2013). A list of novel targets identified with Clone Mapper for the RNAi clones used by Fievet et al. was used as input to GeneMania (yellow circles), together with the genes corresponding to the 14 mutant strains used in the study (black circles). (C) Roy et al. performed a screen to find components of a regulatory network that promotes developmentally programmed cell-cycle quiescence (Roy et al. 2014). Novel targets identified with Clone Mapper for the RNAi clones used by Roy et al. (yellow), together with common targets (black) were used as input to GeneMania. The networks were trimmed to retain only direct neighbors; unconnected genes are not shown. Genes that are linked within GeneMania but do not appear on the list of RNAi clone targets are shown as gray circles; their size is proportional to the calculated probability score. Networks were displayed in Cytoscape; green edges represent experimentally-determined genetic interactions, pink edges represent experimentally-determined physical interactions for the corresponding proteins, orange and gray edges interactions predicted on the basis of co-expression or literature mining, respectively.